













| General Information: |
|
| Name(s) found: |
gi|79313325
[NCBI NR]
gi|42565113 [NCBI NR] gi|28827622 [NCBI NR] gi|28393360 [NCBI NR] gi|11994740 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 761 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
DNA binding
[IEA]
protein dimerization activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSDLEPVAL TPQKQDSAWK HCEVYKYGDR VQMRCLYCRK MFKGGGITRV KEHLAGKKGQ 60
61 GTICDQVPDE VRLFLQQCID GTVRRQRKRR KSSPEPLPIA YFPPCEVETQ VAASSDVNNG 120
121 FKSPSSDVVV GQSTGRTKQR TYRSRKNNAF ERNDLANVEV DRDMDNLIPV AISSVKNIVH 180
181 PTSKEREKTV HMAMGRFLFD IGADFDAANS VNVQPFIDAI VSGGFGVSIP THEDLRGWIL 240
241 KSCVEEVKKE IDECKTLWKR TGCSVLVQEL NSNEGPLILK FLVYCPEKVV FLKSVDASEI 300
301 LDSEDKLYEL LKEVVEEIGD TNVVQVITKC EDHYAAAGKK LMDVYPSLYW VPCAAHCIDK 360
361 MLEEFGKMDW IREIIEQART VTRIIYNHSG VLNLMRKFTF GNDIVQPVCT SSATNFTTMG 420
421 RIADLKPYLQ AMVTSSEWND CSYSKEAGGL AMTETINDED FWKALTLANH ITAPILRVLR 480
481 IVCSERKPAM GYVYAAMYRA KEAIKTNLAH REEYIVYWKI IDRWWLQQPL YAAGFYLNPK 540
541 FFYSIDEEMR SEIHLAVVDC IEKLVPDVNI QDIVIKDINS YKNAVGIFGR NLAIRARDTM 600
601 LPAEWWSTYG ESCLNLSRFA IRILSQTCSS SIGSVRNLTS ISQIYESKNS IERQRLNDLV 660
661 FVQYNMRLRR IGSESSGDDT VDPLSHSNME VLEDWVSRNQ VCIEGNGSSD WKSLEFIKRS 720
721 EEVAVVIDET EDLGSGFDDA EIFKGEKEAS YEGGYSENLF T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
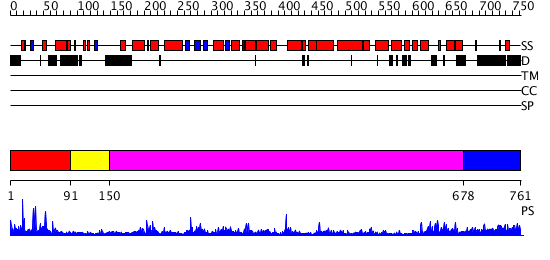
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 1.11 | The structure of the zinc finger from the human spliceosomal protein U1C | |
| 2 | View Details | [91..149] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [150..677] | 37.69897 | Three-dimensional structure of the Hermes DNA transposase | |
| 4 | View Details | [678..761] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)