













| General Information: |
|
| Name(s) found: |
ARC6_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 801 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast inner membrane
[IDA]
chloroplast [IDA] chloroplast envelope [IDA] |
| Biological Process: |
chloroplast fission
[IMP]
protein folding [ISS] chloroplast organization [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEALSHVGIG LSPFQLCRLP PATTKLRRSH NTSTTICSAS KWADRLLSDF NFTSDSSSSS 60
61 FATATTTATL VSPPPSIDRP ERHVPIPIDF YQVLGAQTHF LTDGIRRAFE ARVSKPPQFG 120
121 FSDDALISRR QILQAACETL SNPRSRREYN EGLLDDEEAT VITDVPWDKV PGALCVLQEG 180
181 GETEIVLRVG EALLKERLPK SFKQDVVLVM ALAFLDVSRD AMALDPPDFI TGYEFVEEAL 240
241 KLLQEEGASS LAPDLRAQID ETLEEITPRY VLELLGLPLG DDYAAKRLNG LSGVRNILWS 300
301 VGGGGASALV GGLTREKFMN EAFLRMTAAE QVDLFVATPS NIPAESFEVY EVALALVAQA 360
361 FIGKKPHLLQ DADKQFQQLQ QAKVMAMEIP AMLYDTRNNW EIDFGLERGL CALLIGKVDE 420
421 CRMWLGLDSE DSQYRNPAIV EFVLENSNRD DNDDLPGLCK LLETWLAGVV FPRFRDTKDK 480
481 KFKLGDYYDD PMVLSYLERV EVVQGSPLAA AAAMARIGAE HVKASAMQAL QKVFPSRYTD 540
541 RNSAEPKDVQ ETVFSVDPVG NNVGRDGEPG VFIAEAVRPS ENFETNDYAI RAGVSESSVD 600
601 ETTVEMSVAD MLKEASVKIL AAGVAIGLIS LFSQKYFLKS SSSFQRKDMV SSMESDVATI 660
661 GSVRADDSEA LPRMDARTAE NIVSKWQKIK SLAFGPDHRI EMLPEVLDGR MLKIWTDRAA 720
721 ETAQLGLVYD YTLLKLSVDS VTVSADGTRA LVEATLEESA CLSDLVHPEN NATDVRTYTT 780
781 RYEVFWSKSG WKITEGSVLA S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
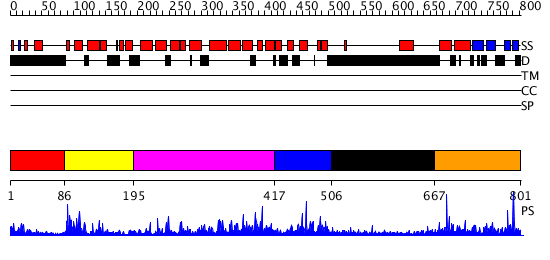
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..194] | 21.39794 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [195..416] | 32.522879 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [417..505] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [506..666] | 3.038997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [667..801] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)