













| General Information: |
|
| Name(s) found: |
SMC6A_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1058 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
membrane [IEA] chromosome [IEA] |
| Biological Process: |
sister chromatid cohesion
[IMP]
response to X-ray [IMP] double-strand break repair via homologous recombination [IMP] chromosome segregation [ISS] |
| Molecular Function: |
two-component sensor activity
[IEA]
ATP binding [IEA][ISS] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEHGDHRQS NPFNDQQTSS GKILRIRLEN FMCHSNLEIE FGDWVNFITG QNGSGKSAIL 60
61 TALCVAFGCR ARGTQRAATL KDFIKTGCSY ALVYVELKNQ GEDAFKPEIY GDTLIIERRI 120
121 SDSTSLTVLK DHQGRKISSR KEELRELVEH YNIDVENPCV IMSQDKSREF LHSGNDKDKF 180
181 KFFYKATLLQ QVDDILQSIG TKLNSANALL DEMEKTIKPI EKEINELLEK IKNMEHVEEI 240
241 TQQVLHLKKK LAWSWVYDVD RQLKEQNEKI VKFKERVPTC QNKIDRKLGE VESLRVSLTE 300
301 KKAQVACLID ESTAMKRELE CLRQSMKKAA REKIALEEEY HHKCSNIQKI KDRVRRLERQ 360
361 IEDINEMTIR STQVEQSEIE GKLNQLTVEV EKAESLVSSL KEEENMVMEK ASAGGKEKEH 420
421 IEEMIRDHEK KQRNMNAHIN DLKKHQTNKV TAFGGDKVIN LLRAIERHHR RFKMPPIGPI 480
481 GAHVTLINGN RWASAVEQAL GNLLNAFIVT DHKDLVALRD CGKEAKYNNL KIIIYDFSRP 540
541 RLDIPRHMIP QTEHPTILSV LHSENTTVLN VLVDVSCVER HVLAENYEVG KIIAFERRLS 600
601 HLKDVFTIDG YRMFSRGPVQ TTLPPRPRRP TRLCASFDDQ IKDLEIEASR EQSEIQECRG 660
661 QKREAEMNLE GLESTMRRLK KQRTQLEKDL TRKELEMQDL KNSVASETKA SPTSSVNELH 720
721 LEIMKFQKEI EEKESLLEKL QDSLKEAELK ANELKASYEN LYESAKGEIE ALEKAEDELK 780
781 EKEDELHSAE TEKNHYEDIM KDKVLPEIKQ AETIYKELEM KRQESNKKAS IICPESEIKA 840
841 LGPWDGPTPL QLSAQINKIN HRLKRENENY SESIDDLRIM HGEKEQKIGK KRKTYKSCRE 900
901 KLKVCKDAVD SRWNKLQRNK DLLKRELTWQ FNHHLGKKGI SGNIRVSYED KTLSIEVKMP 960
961 QDATNSAVRD TRGLSGGERS FSTLCFTLAL QNMTEAPIRA MDEFDVFMDA VSRKISLDTL1020
1021 IDFALKQGSQ WMFITPHDIS MVKSHEKIKK QQMAAPRS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
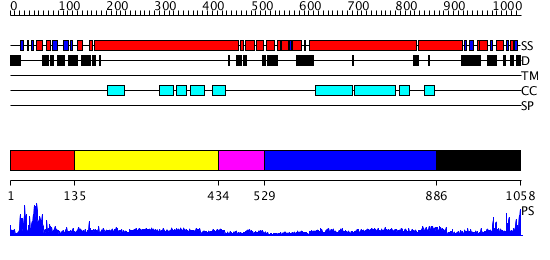
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 35.522879 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [135..433] | 19.09691 | Heavy meromyosin subfragment | |
| 3 | View Details | [434..528] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [529..885] | 10.09691 | Tropomyosin | |
| 5 | View Details | [886..1058] | 22.30103 | Sc Smc1hd:Scc1-C complex, ATPgS |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)