













| General Information: |
|
| Name(s) found: |
RGL1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
|
| Biological Process: |
negative regulation of gibberellic acid mediated signaling pathway
[IMP]
response to gibberellin stimulus [IEP] hyperosmotic salinity response [IGI] response to abscisic acid stimulus [IGI] response to salt stress [IGI] regulation of oxygen and reactive oxygen species metabolic process [IGI] salicylic acid mediated signaling pathway [IGI] gibberellic acid mediated signaling pathway [TAS] jasmonic acid mediated signaling pathway [IGI] response to ethylene stimulus [IGI] |
| Molecular Function: |
transcription factor activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKREHNHRES SAGEGGSSSM TTVIKEEAAG VDELLVVLGY KVRSSDMADV AHKLEQLEMV 60
61 LGDGISNLSD ETVHYNPSDL SGWVESMLSD LDPTRIQEKP DSEYDLRAIP GSAVYPRDEH 120
121 VTRRSKRTRI ESELSSTRSV VVLDSQETGV RLVHALLACA EAVQQNNLKL ADALVKHVGL 180
181 LASSQAGAMR KVATYFAEGL ARRIYRIYPR DDVALSSFSD TLQIHFYESC PYLKFAHFTA 240
241 NQAILEVFAT AEKVHVIDLG LNHGLQWPAL IQALALRPNG PPDFRLTGIG YSLTDIQEVG 300
301 WKLGQLASTI GVNFEFKSIA LNNLSDLKPE MLDIRPGLES VAVNSVFELH RLLAHPGSID 360
361 KFLSTIKSIR PDIMTVVEQE ANHNGTVFLD RFTESLHYYS SLFDSLEGPP SQDRVMSELF 420
421 LGRQILNLVA CEGEDRVERH ETLNQWRNRF GLGGFKPVSI GSNAYKQASM LLALYAGADG 480
481 YNVEENEGCL LLGWQTRPLI ATSAWRINRV E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
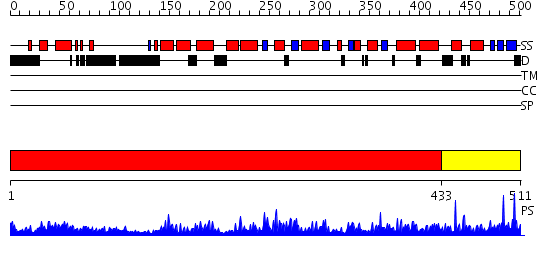
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..432] | 0.971 | Crystal structure of histone K79 methyltransferase Dot1p from yeast | |
| 2 | View Details | [433..511] | 0.988 | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)