













| General Information: |
|
| Name(s) found: |
gi|225879156
[NCBI NR]
gi|15237559 [NCBI NR] gi|10177678 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 557 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
metabolic process
[IEA]
regulation of transcription, DNA-dependent [ISS] |
| Molecular Function: |
DNA binding
[ISS]
zinc ion binding [IEA] protein binding [IEA] N-acetyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSCDIVPME AELSPGAIEE WLSTVKHGKM TENGKRRSDL SIKVKRHLSA LGWVISYYNK 60
61 RNKKEQRYKS PKGKWFYSLA KACMSCVDQD SQQQLQIVPK YDLSCLPSNI ASESSSDVSN 120
121 QKQKKSKKRG RVEDLDVKST APEKDNHPSA DLIKIVPNFD VAKQQQQKKI IMNREGSDTA 180
181 VFNGDQEKIR IGLNVDVIHE QQKKRRKMAS EEIRRPKMVK SLKKVLQVME KKQQKNKHEK 240
241 ESLRFCRKDC SPDMNCDVCC VCHWGGDLLL CDGCPSAFHH ACLGLSSLPE EDLWFCPCCC 300
301 CDICGSMESP ANSKLMACEQ CQRRFHLTCL KEDSCIVSSR GWFCSSQCNR VFSALENLLG 360
361 SKIAVGNDGD LVWTLMRAPN EGEHYDDEQI SKLESAVEIL HQGFEPTNDV FSGRDLVEEL 420
421 IYRKDRTGVG RGFYTVLIER KNEPITVAAV RVDKDVVEIP LVATLSSYRR SGMCRVLMDE 480
481 LEKQMSQMGV CRLVLPAAKE VVTTWTERFG FSVMNSSERL ELVKHGMLDF VGTIMCHKFL 540
541 QKERAENDSA EESSLTE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
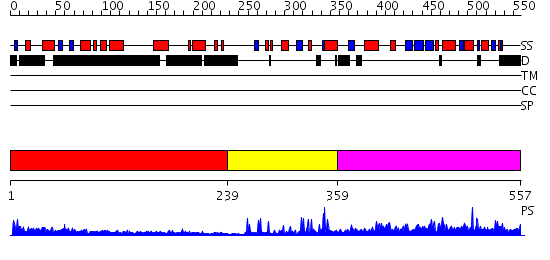
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [239..358] | 12.30103 | No description for 2ysmA was found. | |
| 3 | View Details | [359..557] | 4.33 | Histone acetyltransferase domain of P300/CBP associating factor, PCAF |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)