













| General Information: |
|
| Name(s) found: |
CRD1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 409 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast inner membrane
[IDA]
chloroplast [IDA] chloroplast envelope [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
chlorophyll biosynthetic process
[IEA]
photosynthesis [IEA] |
| Molecular Function: |
DNA binding
[TAS]
magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase activity [IMP][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAEMALVKP ISKFSSPKLS NPSKFLSGRR FSTVIRMSAS SSPPPPTTAT SKSKKGTKKE 60
61 IQESLLTPRF YTTDFEEMEQ LFNTEINKNL NEAEFEALLQ EFKTDYNQTH FVRNKEFKEA 120
121 ADKLQGPLRQ IFVEFLERSC TAEFSGFLLY KELGRRLKKT NPVVAEIFSL MSRDEARHAG 180
181 FLNKGLSDFN LALDLGFLTK ARKYTFFKPK FIFYATYLSE KIGYWRYITI YRHLKENPEF 240
241 QCYPIFKYFE NWCQDENRHG DFFSALMKAQ PQFLNDWQAK LWSRFFCLSV YVTMYLNDCQ 300
301 RTNFYEGIGL NTKEFDMHVI IETNRTTARI FPAVLDVENP EFKRKLDRMV VSYEKLLAIG 360
361 ETDDASFIKT LKRIPLVTSL ASEILAAYLM PPVESGSVDF AEFEPNLVY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
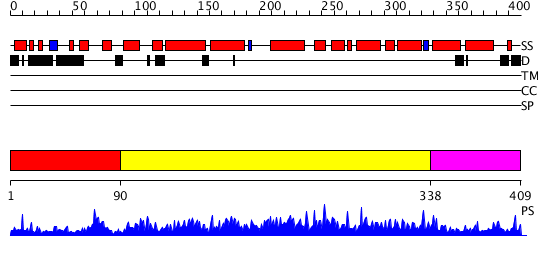
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 1.026983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [90..337] | 0.967 | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | |
| 3 | View Details | [338..409] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)