













| General Information: |
|
| Name(s) found: |
LDL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
histone H3-K4 methylation
[IMP]
|
| Molecular Function: |
oxidoreductase activity
[IEA]
amine oxidase activity [ISS] electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSPASDETA PRRNRRKVSR KNYDENAMDE LIEKQLGGKA KKKYRTKQDL EKETETEALI 60
61 ALSVGFPIDE LLEEEIRAGV VRELGGKEQN DYIVVRNHIV ARWRGNVGIW LLKDQIRETV 120
121 SSDFEHLISA AYDFLLFNGY INFGVSPLFA PYIPEEGTEG SVIVVGAGLA GLAAARQLLS 180
181 FGFKVLVLEG RSRPGGRVYT QKMGGKDRFA AVELGGSVIT GLHANPLGVL ARQLSIPLHK 240
241 VRDNCPLYNS EGVLVDKVAD SNVEFGFNKL LDKVTEVREM MEGAAKKISL GEVLETLRVL 300
301 YGVAKDSEER KLFDWHLANL EYANAGCLSN LSAAYWDQDD PYEMGGDHCF LAGGNWRLIN 360
361 ALAEGLPIIY GKSVDTIKYG DGGVEVISGS QIFQADMILC TVPLGVLKKR SIKFEPELPR 420
421 RKQAAIDRLG FGLLNKVAML FPSVFWGDEL DTFGCLNESS INRGEFFLFY AYHTVSGGPA 480
481 LVALVAGEAA QRFECTEPSV LLHRVLKKLR GIYGPKGVVV PDPIQTVCTR WGSDPLSYGS 540
541 YSHVRVGSSG VDYDILAESV SNRLFFAGEA TTRQHPATMH GAYLSGLREA SKILHVANYL 600
601 RSNLKKPVQR YSGVNINVLE DMFKRPDIAI GKLSFVFNPL TDDPKSFGLV RVCFDNFEED 660
661 PTNRLQLYTI LSREQANKIK ELDENSNESK LSCLMNTLGL KLMGANSVLD TGGALISVIA 720
721 NARRGRSRSH VVAGQCNLPL NPLHFN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
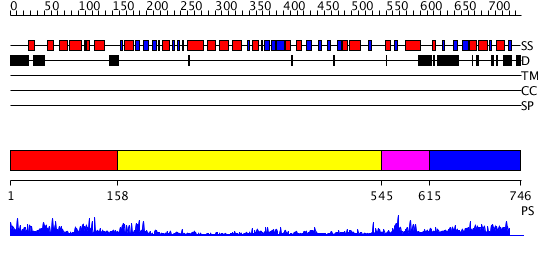
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 27.30103 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 2 | View Details | [158..544] | 15.69897 | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | |
| 3 | View Details | [545..614] | 46.0 | No description for 2v1dA was found. | |
| 4 | View Details | [615..746] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.44 |
Source: Reynolds et al. (2008)