













| General Information: |
|
| Name(s) found: |
PUB44_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 801 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
regulation of chlorophyll catabolic process
[IMP]
regulation of chlorophyll biosynthetic process [IMP] senescence [IMP] regulation of abscisic acid biosynthetic process [IMP] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVGSSDGDQS DDSSHFERGV DHIYEAFICP LTKEVMHDPV TLENGRTFER EAIEKWFKEC 60
61 RDSGRPPSCP LTSQELTSTD VSASIALRNT IEEWRSRNDA AKLDIARQSL FLGNAETDIL 120
121 QALMHVRQIC RTIRSNRHGV RNSQLIHMII DMLKSTSHRV RYKALQTLQV VVEGDDESKA 180
181 IVAEGDTVRT LVKFLSHEPS KGREAAVSLL FELSKSEALC EKIGSIHGAL ILLVGLTSSN 240
241 SENVSIVEKA DRTLENMERS EEIVRQMASY GRLQPLLGKL LEGSPETKLS MASFLGELPL 300
301 NNDVKVLVAQ TVGSSLVDLM RSGDMPQREA ALKALNKISS FEGSAKVLIS KGILPPLIKD 360
361 LFYVGPNNLP IRLKEVSATI LANIVNIGYD FDKATLVSEN RVENLLHLIS NTGPAIQCKL 420
421 LEVLVGLTSC PKTVPKVVYA IKTSGAIISL VQFIEVREND DLRLASIKLL HNLSPFMSEE 480
481 LAKALCGTAG QLGSLVAIIS EKTPITEEQA AAAGLLAELP DRDLGLTQEM LEVGAFEKII 540
541 SKVFGIRQGD IKGMRFVNPF LEGLVRILAR ITFVFNKEAR AINFCREHDV ASLFLHLLQS 600
601 NGQDNIQMVS AMALENLSLE SIKLTRMPDP PPVNYCGSIF SCVRKPHVVN GLCKIHQGIC 660
661 SLRETFCLVE GGAVEKLVAL LDHENVKVVE AALAALSSLL EDGLDVEKGV KILDEADGIR 720
721 HILNVLRENR TERLTRRAVW MVERILRIED IAREVAEEQS LSAALVDAFQ NADFRTRQIA 780
781 ENALKHIDKI PNFSSIFPNI A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
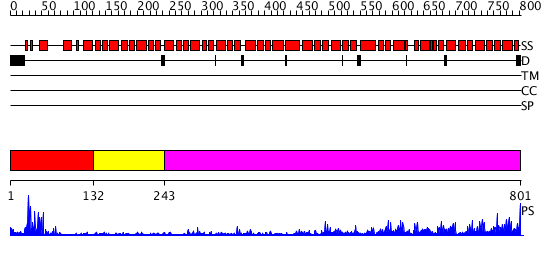
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 6.0 | RAG1 DIMERIZATION DOMAIN | |
| 2 | View Details | [132..242] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [243..801] | 48.522879 | No description for 2z6gA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)