













| General Information: |
|
| Name(s) found: |
RGL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 547 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
|
| Biological Process: |
negative regulation of gibberellic acid mediated signaling pathway
[TAS]
response to gibberellin stimulus [IEP] negative regulation of seed germination [IMP] hyperosmotic salinity response [IGI] response to abscisic acid stimulus [IEP][IGI] response to salt stress [IGI] regulation of oxygen and reactive oxygen species metabolic process [IGI] salicylic acid mediated signaling pathway [IGI] gibberellic acid mediated signaling pathway [TAS] jasmonic acid mediated signaling pathway [IGI] response to ethylene stimulus [IGI] |
| Molecular Function: |
transcription factor activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRGYGETWD PPPKPLPASR SGEGPSMADK KKADDDNNNS NMDDELLAVL GYKVRSSEMA 60
61 EVAQKLEQLE MVLSNDDVGS TVLNDSVHYN PSDLSNWVES MLSELNNPAS SDLDTTRSCV 120
121 DRSEYDLRAI PGLSAFPKEE EVFDEEASSK RIRLGSWCES SDESTRSVVL VDSQETGVRL 180
181 VHALVACAEA IHQENLNLAD ALVKRVGTLA GSQAGAMGKV ATYFAQALAR RIYRDYTAET 240
241 DVCAAVNPSF EEVLEMHFYE SCPYLKFAHF TANQAILEAV TTARRVHVID LGLNQGMQWP 300
301 ALMQALALRP GGPPSFRLTG IGPPQTENSD SLQQLGWKLA QFAQNMGVEF EFKGLAAESL 360
361 SDLEPEMFET RPESETLVVN SVFELHRLLA RSGSIEKLLN TVKAIKPSIV TVVEQEANHN 420
421 GIVFLDRFNE ALHYYSSLFD SLEDSYSLPS QDRVMSEVYL GRQILNVVAA EGSDRVERHE 480
481 TAAQWRIRMK SAGFDPIHLG SSAFKQASML LSLYATGDGY RVEENDGCLM IGWQTRPLIT 540
541 TSAWKLA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
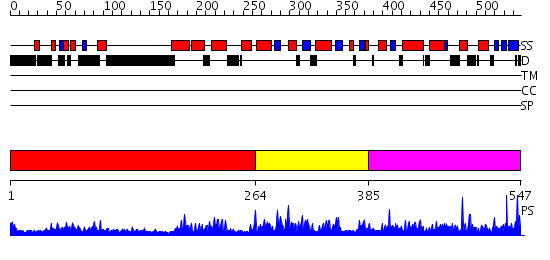
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..263] | 2.076956 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [264..384] | 3.083971 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [385..547] | 3.083954 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)