













| General Information: |
|
| Name(s) found: |
SSY1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 652 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
amylopectin biosynthetic process
[IDA]
|
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
starch synthase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLQISGSV KFEPFVGFNR IRHFRPIASL GFPRFRRRFS IGRSLLLRRS SSFSGDSRES 60
61 DEERFITDAE RDGSGSVLGF QLTPPGDQQT VSTSTGEITH HEEKKEAIDQ IVMADFGVPG 120
121 NRAVEEGAAE VGIPSGKAEV VNNLVFVTSE AAPYSKTGGL GDVCGSLPIA LAGRGHRVMV 180
181 ISPRYLNGTA ADKNYARAKD LGIRVTVNCF GGSQEVSFYH EYRDGVDWVF VDHKSYHRPG 240
241 NPYGDSKGAF GDNQFRFTLL CHAACEAPLV LPLGGFTYGE KSLFLVNDWH AGLVPILLAA 300
301 KYRPYGVYKD ARSILIIHNL AHQGVEPAAT YTNLGLPSEW YGAVGWVFPT WARTHALDTG 360
361 EAVNVLKGAI VTSDRIITVS QGYAWEITTV EGGYGLQDLL SSRKSVINGI TNGINVDEWN 420
421 PSTDEHIPFH YSADDVSEKI KCKMALQKEL GLPIRPECPM IGFIGRLDYQ KGIDLIQTAG 480
481 PDLMVDDIQF VMLGSGDPKY ESWMRSMEET YRDKFRGWVG FNVPISHRIT AGCDILLMPS 540
541 RFEPCGLNQL YAMRYGTIPV VHGTGGLRDT VENFNPYAEG GAGTGTGWVF TPLSKDSMVS 600
601 ALRLAAATYR EYKQSWEGLM RRGMTRNYSW ENAAVQYEQV FQWVFMDPPY VS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
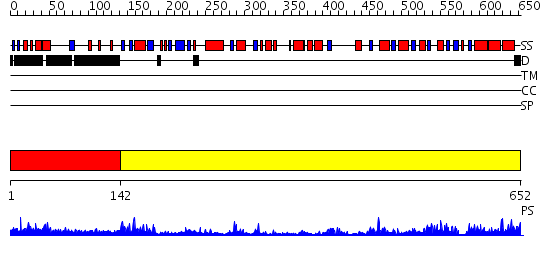
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..652] | 77.30103 | Crystal structure of the glycogen synthase from A. tumefaciens in complex with ADP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)