













| General Information: |
|
| Name(s) found: |
PLDA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
chloroplast [IDA] membrane [IDA] mitochondrion [IDA] clathrin-coated vesicle [IDA] plasma membrane [IDA] |
| Biological Process: |
regulation of stomatal movement
[IMP]
response to cadmium ion [IEP] seed germination [IMP] fatty acid metabolic process [IMP][TAS] positive regulation of abscisic acid mediated signaling pathway [IMP] |
| Molecular Function: |
phospholipase D activity
[IDA][IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQHLLHGTL HATIYEVDAL HGGGVRQGFL GKILANVEET IGVGKGETQL YATIDLQKAR 60
61 VGRTRKIKNE PKNPKWYESF HIYCAHLASD IIFTVKDDNP IGATLIGRAY IPVDQVINGE 120
121 EVDQWVEILD NDRNPIQGGS KIHVKLQYFH VEEDRNWNMG IKSAKFPGVP YTFFSQRQGC 180
181 KVSLYQDAHI PDNFVPRIPL AGGKNYEPQR CWEDIFDAIS NAKHLIYITG WSVYAEIALV 240
241 RDSRRPKPGG DVTIGELLKK KASEGVRVLL LVWDDRTSVD VLKKDGLMAT HDEETENFFR 300
301 GSDVHCILCP RNPDDGGSIV QSLQISTMFT HHQKIVVVDS EMPSRGGSEM RRIVSFVGGI 360
361 DLCDGRYDTP FHSLFRTLDT VHHDDFHQPN FTGAAITKGG PREPWHDIHS RLEGPIAWDV 420
421 MYNFEQRWSK QGGKDILVKL RDLSDIIITP SPVMFQEDHD VWNVQLFRSI DGGAAAGFPE 480
481 SPEAAAEAGL VSGKDNIIDR SIQDAYIHAI RRAKDFIYVE NQYFLGSSFA WAADGITPED 540
541 INALHLIPKE LSLKIVSKIE KGEKFRVYVV VPMWPEGLPE SGSVQAILDW QRRTMEMMYK 600
601 DVIQALRAQG LEEDPRNYLT FFCLGNREVK KDGEYEPAEK PDPDTDYMRA QEARRFMIYV 660
661 HTKMMIVDDE YIIIGSANIN QRSMDGARDS EIAMGGYQPH HLSHRQPARG QIHGFRMSLW 720
721 YEHLGMLDET FLDPSSLECI EKVNRISDKY WDFYSSESLE HDLPGHLLRY PIGVASEGDI 780
781 TELPGFEFFP DTKARILGTK SDYLPPILTT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
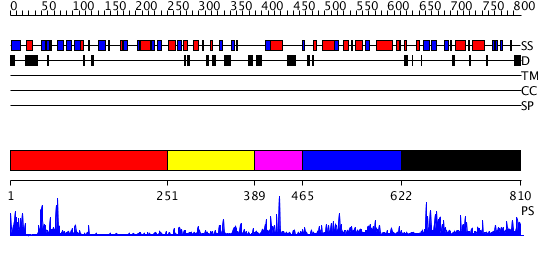
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | 19.0 | Synaptogamin I | |
| 2 | View Details | [251..388] | 5.39794 | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | |
| 3 | View Details | [389..464] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [465..621] | 6.09691 | Nuclease Nuc | |
| 5 | View Details | [622..810] | 1.338976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)