













| General Information: |
|
| Name(s) found: |
GCSP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1044 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] chloroplast envelope [IDA] glycine cleavage complex [ISS] |
| Biological Process: |
glycine decarboxylation via glycine cleavage system
[ISS]
glycine metabolic process [IEA] |
| Molecular Function: |
ATP binding
[IDA]
glycine dehydrogenase (decarboxylating) activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERARRLAYR GIVKRLVNET KRHRNGESSL LPTTTVTPSR YVSSVSSFLH RRRDVSGSAF 60
61 TTSGRNQHQT RSISVDALKP SDTFPRRHNS ATPDEQAQMA NYCGFDNLNT LIDSTVPKSI 120
121 RLDSMKFSGI FDEGLTESQM IEHMSDLASK NKVFKSFIGM GYYNTHVPPV ILRNIMENPA 180
181 WYTQYTPYQA EISQGRLESL LNYQTVITDL TGLPMSNASL LDEGTAAAEA MAMCNNILKG 240
241 KKKTFVIASN CHPQTIDVCK TRADGFDLKV VTVDIKDVDY SSGDVCGVLV QYPGTEGEVL 300
301 DYGEFVKNAH ANGVKVVMAT DLLALTMLKP PGEFGADIVV GSGQRFGVPM GYGGPHAAFL 360
361 ATSQEYKRMM PGRIIGVSVD SSGKQALRMA MQTREQHIRR DKATSNICTA QALLANMTAM 420
421 YAVYHGPEGL KSIAQRVHGL AGVFALGLKK LGTAQVQDLP FFDTVKVTCS DATAIFDVAA 480
481 KKEINLRLVD SNTITVAFDE TTTLDDVDKL FEVFASGKPV QFTAESLAPE FNNAIPSSLT 540
541 RESPYLTHPI FNMYHTEHEL LRYIHKLQNK DLSLCHSMIP LGSCTMKLNA TTEMMPVTWP 600
601 SFTNMHPFAP VEQAQGYQEM FTNLGELLCT ITGFDSFSLQ PNAGAAGEYA GLMVIRAYHM 660
661 SRGDHHRNVC IIPVSAHGTN PASAAMCGMK IVAVGTDAKG NINIEELRNA AEANKDNLAA 720
721 LMVTYPSTHG VYEEGIDEIC NIIHENGGQV YMDGANMNAQ VGLTSPGFIG ADVCHLNLHK 780
781 TFCIPHGGGG PGMGPIGVKQ HLAPFLPSHP VIPTGGIPEP EQTSPLGTIS AAPWGSALIL 840
841 PISYTYIAMM GSGGLTDASK IAILNANYMA KRLESHYPVL FRGVNGTVAH EFIIDLRGFK 900
901 NTAGIEPEDV AKRLMDYGFH GPTMSWPVPG TLMIEPTESE SKAELDRFCD ALISIREEIS 960
961 QIEKGNADPN NNVLKGAPHP PSLLMADTWK KPYSREYAAF PAPWLRSSKF WPTTGRVDNV1020
1021 YGDRNLVCTL QPANEEQAAA AVSA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
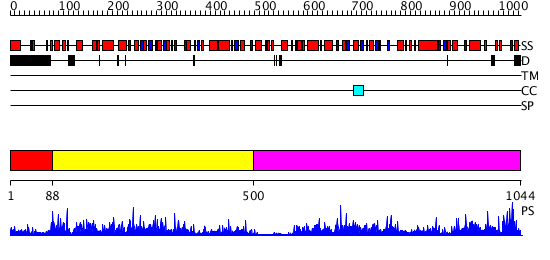
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..499] | 77.69897 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | |
| 3 | View Details | [500..1044] | 82.0 | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)