













| General Information: |
|
| Name(s) found: |
ATK3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 754 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule associated complex
[IEA]
|
| Biological Process: |
microtubule-based movement
[IEA]
|
| Molecular Function: |
ATPase activity
[IDA]
microtubule motor activity [ISS] microtubule binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVGAMANNGR IRSAFPVTNG SKDLTPNSAP ASTTGSEYGP VEFTREDVET LLNERIKYKS 60
61 KFNYKERCEN MMDYIKRLRL CIRWFQELEL DYAFEQEKLK NALELNEKHC VDMEVSLKNK 120
121 EEELNMIIEE LRKNFESVQV QLAREQTEKL AANDSLGKEK EARLSVEKAQ AGLTEELGKA 180
181 QGDLQTANQR IQSVNDMYKL LQEYNSSLQL YNSKLQGDLD EAHETIKRGE KERTAIIENI 240
241 GNLKGQFSAL QEQLAASKAS QEDIMKQKGE LVNEIASLKV ELQQVKDDRD RHLVEVKTLQ 300
301 TEATKYNDFK DAITELETTC SSQSTQIRQL QDRLVNSERR LQVSDLSTFE KMNEYEDQKQ 360
361 SIIDLKSRVE EAELKLVEGE KLRKKLHNTI LELKGNIRVF CRVRPLLPGE NNGDEGKTIS 420
421 YPTSLEALGR GIDLMQNAQK HAFTFDKVFA PTASQEDVFT EISQLVQSAL DGYKVCIFAY 480
481 GQTGSGKTYT MMGRPGNVEE KGLIPRCLEQ IFETRQSLRS QGWKYELQVS MLEIYNETIR 540
541 DLLSTNKEAV RTDSGVSPQK HAIKHDASGN THVAELTILD VKSSREVSFL LDHAARNRSV 600
601 GKTQMNEQSS RSHFVFTLRI SGVNESTEQQ VQGVLNLIDL AGSERLSKSG STGDRLKETQ 660
661 AINKSLSSLG DVIFALAKKE DHVPFRNSKL TYLLQPCLGG DAKTLMFVNI APESSSTGES 720
721 LCSLRFAARV NACEIGTPRR QTNIKPLENR LSLG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
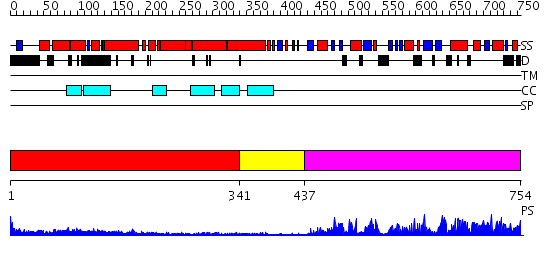
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..340] | 18.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [341..436] | 1.32 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 3 | View Details | [437..754] | 93.39794 | No description for 2repA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)