













| General Information: |
|
| Name(s) found: |
zen-4 /
CE12346
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 772 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cleavage furrow
[IDA microtubule associated complex [IEA |
| Biological Process: |
embryonic cleavage
[IMP embryonic development ending in birth or egg hatching [IMP cytokinesis [IMP microtubule-based movement [IEA |
| Molecular Function: |
ATP binding
[IEA microtubule motor activity [IEA plus-end-directed microtubule motor activity [ISS microtubule binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSRKRGITP SRDQVRRKKL SIEETDSIEV VCRLCPYTGS TPSLIAIDEG SIQTVLPPAQ 60
61 FRRENAPQVE KVFRFGRVFS ENDGQATVFE RTSVDLILNL LKGQNSLLFT YGVTGSGKTY 120
121 TMTGKPTETG TGLLPRTLDV IFNSINNRVE KCIFYPSALN TFEIRATLDA HLKRHQMAAD 180
181 RLSTSREITD RYCEAIKLSG YNDDMVCSVF VTYVEIYNNY CYDLLEDARN GVLTKREIRH 240
241 DRQQQMYVDG AKDVEVSSSE EALEVFCLGE ERRRVSSTLL NKDSSRSHSV FTIKLVMAPR 300
301 AYETKSVYPT MDSSQIIVSQ LCLVDLAGSE RAKRTQNVGE RLAEANSINQ SLMTLRQCIE 360
361 VLRRNQKSSS QNLEQVPYRQ SKLTHLFKNY LEGNGKIRMV ICVNPKPDDY DENMSALAFA 420
421 EESQTIEVKK QVERMPSERI PHSFFTQWNS ELDGSVRMED DGSREIPCPP TFCLTDCNDK 480
481 DTVDSMYKYA RKLSSLQNSS EEGPSSTLLT MIRQYMMEAD YQRVEIARLK DSLNDKDEEI 540
541 KKLRGFCSRY KRENASMKER IASCEQGEQE NALVMEKLME QKMEDRKIIQ SQKKAMRNVR 600
601 GIIDNPSPSV ASLRSRFDQE NVAHPTAPIQ TPPPPYQTPG RAPVFKKRLE ATTSTTVMSG 660
661 SSSGGSGQQG YVNPKYQRRS KSASRLLDHQ PLHRVPTGTV LQSRTPANAI RTTKPEMHQL 720
721 NKSGEYRLTH QEVDDEGNIS TNIVKVNSLV STQKHACTVP LSFSRVLITH LS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
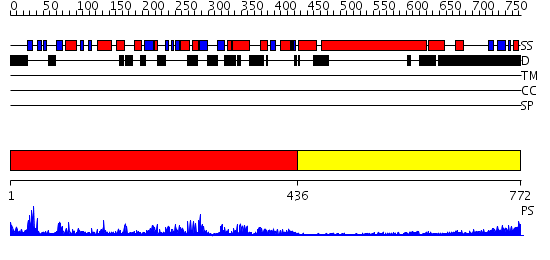
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..435] | 77.221849 | Kif1a head-microtubule complex | |
| 2 | View Details | [436..772] | 5.154902 | No description for 2dfsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)