













| General Information: |
|
| Name(s) found: |
SPAC23H4.15
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS nucleolus [ISS |
| Biological Process: |
ribosome biogenesis
[ISS rRNA processing [ISS |
| Molecular Function: |
ribonucleoprotein binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHHHRSTFK AKKPFKSKHA SKSSLKEKYK NEVEPHRSGP KNIVHTSTKA DRRNTAKQIQ 60
61 LNKRTEVAMN NRIFGGKNGA PKVITIVPLC NNVDSWNVLT NLLRSIDPEA SLPKFDKDSI 120
121 SYSTTIDRFK QNLLFLLPKR EFYSLIDACK VSDYVIFVLS AVQEVDEFGE LIVRTTQGQG 180
181 ISSVLSMVHD LSEVDSLKTR NEVKKSLQSF MNFFFSDQER VFAADVSQDA LNVMRALCTS 240
241 HPRGIHWRDS RSYLLSQEIS YSNGNLLVRG IVRGKGLDPN RLIHIQGFGD FAINRIYEAP 300
301 QGIQNSRGIS MDEDTNLTGG LVELCSPTQE QDSLESLGPI IDDMDTDMDS EVGKEASRGV 360
361 RLDDFYYFDD EEEPVAVAKR VPKGTSTYQA TWIPDEDEES DQYSDVEDTE VIIEDQDNQE 420
421 ISNHVAEEKI DSDEEETIDD AKSEMFVDLS EEEEVRQYEE YRKKQKELQE ELEFPDEVEL 480
481 QPNELARERF KKYRGLRSLY TSQWDADEYD PNEPREWRQL FKFENYRNLK NKFLKQPFIG 540
541 EAKPGKAVYV ELRNVPIEIF EYYNKPWNLL VLYSLLQYEN KLTVSQFTAM QHSEYEEPIE 600
601 SKEELLLQIG PRRFMVRPLY SDPTASGASN NLQKYHRYLP PKQAVIASVI SPIVFGNVPI 660
661 IMFKKSSDNS LRLAATGSYV NCDTNSVIAK RAVLTGHPFK VHKKLVTIRY MFFNPEDVIW 720
721 FKPIQLFTKQ GRTGYIKEPL GTHGYFKATF NGKITVQDTV AMSLYKRMYP LPCELFKVTD 780
781 IDS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
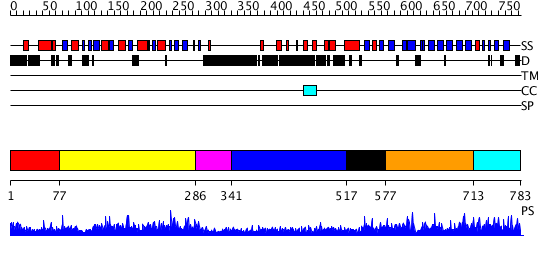
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..285] | 4.30103 | Initiation factor eIF2 gamma subunit, domain II; Initiation factor eIF2 gamma subunit; Initiation factor eIF2 gamma subunit, N-terminal (G) domain | |
| 3 | View Details | [286..340] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [341..516] | 1.06699 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [517..576] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [577..712] | 1.04197 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [713..783] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)