













| General Information: |
|
| Name(s) found: |
snf5 /
SPAC2F7.08c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[IDA nuclear chromatin [IC] cytoplasm [IDA Golgi apparatus [IDA |
| Biological Process: |
chromatin remodeling
[ISS positive regulation of cell-substrate adhesion [IMP positive regulation of gene-specific transcription from RNA polymerase II promoter [IEP negative regulation of gene-specific transcription from RNA polymerase II promoter [IEP regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
general RNA polymerase II transcription factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKDIGSAVA ELQVCSTHYA MTIVSCGFHS PQSLMNADLQ HCLLADQNYL LNNSNKNKGS 60
61 IIEENTRKPF QSARNLQTSL KTHPESAITR YGLGYAGFGN GWTALKPKIL FPSQKRRVKG 120
121 RFTFPFSRRS NLAQSKLPVD LVPIRLEIDA DRYKLRDSFT WNLYDKCISL DQFAEQICID 180
181 YDIPLHNVHI VQNISKSIQA QINDYEPRKA QSNLSFVSDV SSSTSETVYA HEPSDSLAKA 240
241 SKQQIPTVQN DLRILIKLDI TIGRLNLIDQ FEWNLFAPES SAEEFATVMC LDLGLSGEFC 300
301 TAVAHSIREQ CQMYIKYLSL IGYLFDGSEI EDEEVRSYIL PPLKNTLRFS DMESSSFAPM 360
361 IYELNDAEME RQDRGYDREA RRMRRRQGRA KHGIMLPDLR DIPKIHRSLF PSSSVPSDDD 420
421 FMHALSLSTN SDGETLNEIN TNNPEREHLI VRLKVDSQKL KIIVEQSQTL TLNNFQQRKL 480
481 TPLPNSMSFD SLNEFESERN TPSTYGQNIS PLPQFSSPSL SSDAFLTNSN SSESALVHMQ 540
541 KLNLPDFTPS WVKRCVIETF AKFPNDRFNV IVKPALNPAE RMTVRICCHD CINEYFTAGP 600
601 GFTFGNFHIH LLSSSHQQKK EVRMNTVLDR NT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
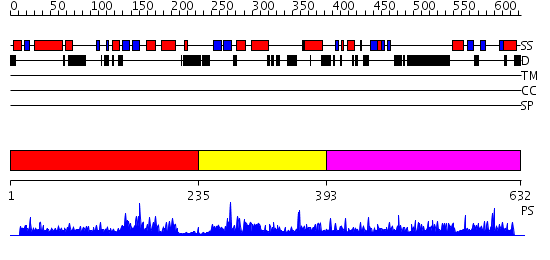
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [235..392] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [393..632] | 40.185997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)