













| General Information: |
|
| Name(s) found: |
px-PA /
FBpp0071749
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1995 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoplasm
[NAS]
|
| Biological Process: |
imaginal disc-derived wing vein morphogenesis
[TAS]
inter-male aggressive behavior [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKAKAYIKN ELRGGSQQSE PATTPNPTSP ASAPGTPQAT RHSYPPVELM PSPGAAAHRS 60
61 ARTPGEQYHF GYQAHPAQHY HSGAKSTGRA SAPPDNHHVP MTICFVCGGH GSFEPQPLRI 120
121 RHNAERPSES YFPFLERHEP PNGVPGVVPG QEYVWACMLC FRSLNEQWDA YERQKKPLLQ 180
181 RIYHMKRVDG KGYIGADVAT QSEYAAQVLG LSAEHLTQNG LVEGHGDPYA LARHYSPQQP 240
241 SHHHHSTSRN ASPSPNPIAA VGGGGEFYPR NSTPSSNHNR YLSRPQSRDM PVASPTSRPP 300
301 SEPPAMKSAG YAQQKFKLSP APPVGYQGQA PYHPAQYQQL QQQTAAYHLL QQQVNAGGGA 360
361 TESAHYKGNP YGALPSPGSG PLGPRAASTL EDDNATVLDL RNSSNSSAPP APRHVGSTAS 420
421 SVSQPSQPQS EVGILDLSMP DKNSITEVCY VCGDEQRRGS LIELSTVKPR ETGRPGPSVP 480
481 YFPLFNEQHP RPARSRPKDP RGMIQACLPC YEDLMQQWNV HQASHKPESE RSYLLRKRST 540
541 PAAERTTFVC YTCGTDTPSS QLRLVYCCPN AEREPYYPFI KTMKVYKNAS PISPQGMVQV 600
601 CSTCNEKHSG LAEGGPPPAS SSIHSAVGGG SPSAGHSLES NSAMYSTSGA ITGNSGAQFG 660
661 GRNSPSEKSL ANSDSMSNVR FRPYETNSNN SNSNTARDLK QIRRSDSRPN SPSSSQGAIE 720
721 NGHGQYPCSI CKVLCPSNKM DWLSTSAEHM NSHAMHFPCL KANAVTNNNN NNNNTNNNNN 780
781 NNNNELNSNR VLACKDCKDY LASQWETMDA ERVPLEHRRY NIPSPMLTSS SPNGSRHGAM 840
841 SINTPPSTPS ISSSTPASTS IYCYLCGLHS DLTLARVLYA SKEGSRPYFP LLLKHNSLPN 900
901 AEQLRNNSAL VCTFCYHSML NQWRKYEAQS PSVNPADRKY NWHDYNCHLC GITTYRKRVR 960
961 ALPVNEFPFV KHQKSDDGLL LENGEYAVVC LDCYESLRQQ ASQHDRFGVP ISRRAYNWVP1020
1021 QPPPPEDSPE SAVARLPCGE RSDRIHINNA LRPVPGKKTT SPKQYDKSKE VPPKAGQKRP1080
1081 ATSPAPHIPV PHHLQTPPGG GAGGLPGSSV ANSPIPSSAG GNALMNHALP NNHIGTPSPQ1140
1141 QQHQHQQHQQ QQQQQQHQAQ QQAQQQAVAA AVQQQLAAGV GLPPGAIGAA SASSRGSFAA1200
1201 ALRTLAKQAD IKEEDVGGDR SVPTSGAGGP PPSSSGPGSL NSVAGRVGSE RPGEDRVSSN1260
1261 KKRSPPSPQP PEKIARLSHT PQTASLQPEL LARSGFQPYR SDERLMHPAG AFPLEAYSHF1320
1321 AGLQGIPPAF LNPAGLPYTE QLYLEHRYQL FRAAGAHPSH PHAAALYPQM ASPYSHLYSM1380
1381 MPGAALGISP AMHDRMKLEE EHRARLAREE EREREIQREK ERELREQREK EQREKEQREK1440
1441 EQREREQREK EQREKEARER KLRDEERERE RERQQLLTAS HHYSNQLYSP LNRNLLGSMI1500
1501 PHLNMSLRGP PGALHGLPGM SHYHAAAANA QRQSPHGAMG LNLGMAGLPG VPPLGGHGPN1560
1561 LQHHLQQAAM GLAHPGAAGL THPGFSAAAL GLGAHPHSLN LSHPHMSPHH PASLAHQLPP1620
1621 HTSSAGGGGL SNSIPVTASS SLSASSPHTS LNLTSSVKLS ADQVPTSTPS GGVSGSGMPP1680
1681 TTSSSHIPNM GHYYHHGHLA AMHAAAAAAS GMTPTAAHQQ QPLSLPSSPK ITPPITPSSS1740
1741 ANGGRAGGSP HAMRHHIAAA VAAAAQQSAM IDKAATPVTH EPTTLDLTGS NSNSSNQAPS1800
1801 NASSGQPTNH EVNGAESNGA SQTESKEHLA IKEDGHKSSP PPVALEKKPA TPNAAADKPI1860
1861 HPDSSPENSS RRSASPQSDA NNSLPAVSVT AAASGKCKEE TPKILDSDTT ATTKTDIEEE1920
1921 KQRARLSVSP TPTAPKIMVA SPDTVSTSGG VTSTTSGTTP PPVVSSSTSS AGKAPALTSD1980
1981 EVSTSAATSA AEPTR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
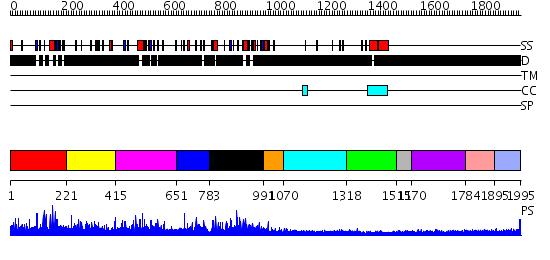
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [221..414] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [415..650] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [651..782] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [783..990] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [991..1069] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1070..1317] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1318..1510] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1511..1569] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1570..1783] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1784..1894] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1895..1995] | 2.208984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.58 |
Source: Reynolds et al. (2008)