













| General Information: |
|
| Name(s) found: |
sha-PA /
FBpp0087221
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1626 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell cortex
[IDA]
|
| Biological Process: |
epidermal cell differentiation
[IMP]
non-sensory hair organization [IMP] antennal morphogenesis [IMP] imaginal disc-derived wing hair organization [IMP] cuticle pattern formation [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLQSLPLLL LLAVVTLPPA TPTSAARPLT HPRARNHYPS EDLAEEPLMP ASLLLEELQQ 60
61 TNGQLVNLTR QHDGDTFHTS GLSSACNADT CIGLSSGTAS LVRSTGDGSG QGNDGLNPAT 120
121 SSLGGGQMEL LTRRLKLQKG RGQVANIDAS GETGCTCRCL PYQRAYREDL GICVDDIHEC 180
181 SLSPFVSGSS SEKIPFVFLP LKGQIIYPSR EISFASIHTP VCAVTGAQYL GSNGWSDLRN 240
241 PIDTDYPFRM FRDEGRSFLL WLGEADLRQK MQGRLIVVHL VCRDMTVALN ASNHVKGEPL 300
301 MPPRNVHSPC VAFRVNGSPV KYAHNVPEVF FQPANSTTLA STSDGMTMRE YVVIGICSLL 360
361 LGLIYVASVF LYLYMKKRKR HSSRHSLDNL TNDINYPKND QVTYGAPFSR VGSIYSSNSM 420
421 GLGNGNESNT RTSIGSLKED MGIVKNNPLL QHFPQLSERE HNSGFASDIS NSNSECEMEE 480
481 KIKTSVLVHP QLSKSPKPSG GTDNAAMDVD DFLQDNECLP AENVAVIDEL MSEEKLENLR 540
541 AMVSGNVRKK LYFNPAYFEP HLLAEPPQAA IEFLQKIREV IAIAKYKMAS KRYQPSLNII 600
601 PEEQHPESSS SSPMFNIPLQ QSLAGKGMGD KRNSIEQWLQ SVPNSDATPA PKTLDRPAPP 660
661 TAKKPSSGSP NKGSLRKEIS APKERPPPPP MQKSKADEGN AKSSSPAPSG PHSSRSTGKS 720
721 SPEQSPKPKT NVGSTYEGFS AFSVAANVYG FAETGGEIYN NPKFMLADSP AASLRSASSR 780
781 SKSMKKRSEV LDQFAAANST PTSLTSFDRQ HYNMLRNMKA PEVIYDSLKR EFAAHIPTPD 840
841 YDSTIEKRIK DIYSSTQSSI PSVPTPDYSS LSRKSLKQFQ PDSPIYRRKS PQYLIVDYET 900
901 DSLERLEPSK RKSSHSSSSP SSDVSSQLSP SLSTALPLEE EMEISHTVYD RMNGFRKEGE 960
961 MKKRLAAKHQ HPHPGQQQRQ KEAAARIKYD TPFRGSMTIE VEHEPPSDLE RTDSDQFEPD1020
1021 TLDRKPKKQS FCDISAWDGH KPIDSDFSQI SSLPDLSRQI SPAPNQGIKP RTASFILRSS1080
1081 NSFRNLREIY ESPLIAQDNI KYEKKVAQTP EEEGRILTLE AKHSRRQRLM ETSPTHSVMD1140
1141 ITKVNKTTIT ISSPKPADKA PPTRPKVNQH YCPPLVQSKR PLPPPPPNSS EHQAPDEDAY1200
1201 SLHSEITGTS EFENSCNNTM SSAISATTEE RTHAKGSRSP KINAKIPEED YEGMYNKKIN1260
1261 SLRNDYSLTK YLNRRKSQRE EVSKAGPDSP KDVAPEPEFG GSNKSVKEVD VAQFDALSIS1320
1321 SRSNRSSSHL SERTKEMYRN AGVPVGIAYP QRAASSSGNA SAGGNVQTVY RCEIEQAQLT1380
1381 AGMQIAIGLK DRAKKAKDLK NAWRKFVTMA ANKFSPSQSP APTYGSKTAA GFLEDDGIAS1440
1441 IIEDAAQSAG MGQPGSQTYY EQRFGQLDSG YMSTDSTELY KRNVYDRYNF KMPCGRGQII1500
1501 RVDTIEDNEE ESGPNDNRFE DLEHMVSPSH SRRSPENNAV VACDLSDAES DKTLTRSHSQ1560
1561 TNSQERNVRG STTTMSSYSS DENGRGRSTC CGSSETDEET NAEDMWESGA ESIETHSVLY1620
1621 KMIRKS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
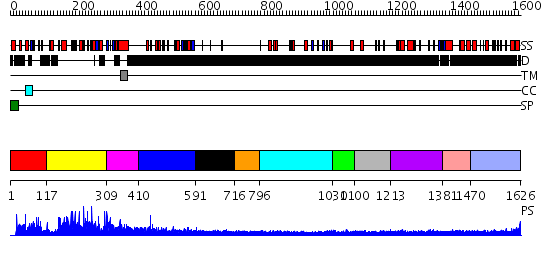
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..308] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [309..409] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [410..590] | 1.229993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [591..715] | 3.249996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [716..795] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [796..1029] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1030..1099] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1100..1212] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1213..1380] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1381..1469] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1470..1626] | 1.178997 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|