













| General Information: |
|
| Name(s) found: |
SFC3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1339 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSLIRHCSE EIALDGSSGC SIDRLWEFAA NFFFRQGIVQ NLDDNYKTFV WPLILKEDGI 60
61 EVWIEDEDAT GLRSKMQEIP WNYQDITIEL RARIRLFASE DRQWLTLTYK TKTDSKIQPL 120
121 AFELLSCISR YRQEGVDRIQ LCKETKQEPR SVYGRIQALE DASLISKVAI NRSRAQTALL 180
181 VLKRFESENS TINETVAQVS GKQIYNTEKI RSNICDAVQS SRNGICRHVD ARAMVNLNHS 240
241 RWERRYYARQ VTYLHNNGYL RKCLTFVPNS PERCIRCLQF IKPYISSLDA TEDDVDFTEV 300
301 DDIPEDDELE EEEPLLPSKE EFDASFLQPL ADVGPTLPQW SRFRPLEFQC FVLIRNAGFH 360
361 GVITLQILSG LTGIRFNKPL FKLLGSLVEH RSSVPNHLSH MSITRFEDKT KKSRQYRYFT 420
421 LQSQLNRLIR DGIPAETISK LLPHVHSLAG EFSPIDSSLF LNSLHHKGNM ESNAEVSPDG 480
481 MTLLPRKRGR PRKSANISVT SSPIRPSKNE NNLPSLAISP VSEGFIQNAT ISTPSTSSNL 540
541 SIAGSLTPSK TSRIYRGLAP LKNPEFNEDH VKKAEHLEPL LNVSSSVPSK NFTVSSPDHL 600
601 KYGNTSSLQV SDSQSSIDLD SSFHYPVSVD SQLSHSTGSL MANFSSPTKK RRLESVDFIF 660
661 LQRKGLILSY LVEQNGAFEI SRKMFEDLAD LKVRRNPETS RTVMDRRTFQ QTLEKLMQEK 720
721 KVRKLVIATN NGLGKLVRKD IVVQYDMKPD SPRFQQLRAQ ITTPEIEKQS TPEILKDVDV 780
781 DFLKRNTSLS RRKSMPAEIK RHKESSETKP VDKEEVKKNE KEKDDPMRLA QQLLESLAPD 840
841 FALHENTQQK SPVEKPKKLR KDRYASVEEF DYFSSTEHAS KRSVKRFKND FTSDEDETLI 900
901 RAVVITQIYY GGTNRLIKWE AVQKCFPNRD IYALTRRYLS IRQHTKFKGL QQFLSENWQQ 960
961 MYKDAVSRKD LMPYPDSVDD FDPTPYVKAS CRPYMISSSN LATTRLPRDL VDVYETYNIE1020
1021 VVKQETNFRE MIFDPSLSVA SKMNAYCDMP FTMPLSLNDK QNNEGDENCE KGQLFDAKST1080
1081 IKSIVAIPDA TYDARFSQER LMQYPEDILI AAHQELLDKR IITRVNSENS RLQPGRNFQF1140
1141 TEKFASSLKS PLPPFLLSQA KRFNKFLLEG FQNHKNHLFE ETSNSGTLAC ILDLLSQGKL1200
1201 QISIVGSKFN EYGLSEGYRT RLLEHDNINV TLVLSGKESE SKKNYGVTEK LATPPSHEPR1260
1261 LWLNGKCELI EMIWMNIEQS IVYQLLRKPG ILRSQLTNLL FPGLEPREFN EVLDYFIAAG1320
1321 AAIEKDGLYL NHNYLFKLT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
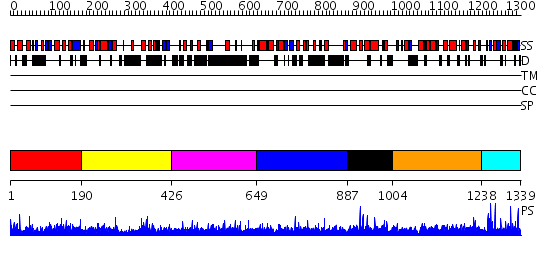
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | 1.036977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [190..425] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [426..648] | 2.113996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [649..886] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [887..1003] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1004..1237] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1238..1339] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)