













| General Information: |
|
| Name(s) found: |
URB2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1318 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLQTLTKRI RDKNGSREDQ IDLANKLFRG EVDVYFPNKE EFIINFLLDR LKENSNLTCV 60
61 NIGYWQLFLN IWKSDKLSGS FRVSAIRRQS LYPIILNAFE YLIKEFKKDV FLKISDVFVC 120
121 VYSNLNILYI SRSFPDQISL LFSRYCYLIA SKIDDLLDSH PNEVFKETLI CVIQSAVHSQ 180
181 ASSTKAQTQF LHQTLGSILA LLCKTQKGDP YFSFLNAVLM KCVLTQHTLD VLLLTKKDSS 240
241 MTLCSLLNAI SFEPINLKAI PLFFIICADS INFHLSSVEK SISQQKASEL MYKLFSEHIA 300
301 FLLSLKNLPE EATLDCMLSI CQVFENDNLH RGHSDEYTFA MLEQLLELSV KTLRQNTCWS 360
361 TSWEMLSSLL AIDFDVLLPH SKILWECIKS VSHQDDIHAL KFLKRWVRAS AKARILDSFC 420
421 IDWHLNVQTM SERSILTNRD FITSFSAVAE VSLSQISIKK LVTWLMSSES IMSENSNRFV 480
481 VYYSLCKSLK RESYPAHLVP NFELFTRSIF NASLKNFNIL DERSQALLCM CQISHVQFFS 540
541 SDMSISASVG MKTLFEDLMA EKSRGLSSTV WSLQLLLCYL EQCLRLNTLH EDTTFPMRVV 600
601 NYALEYSSKA LDKFAENSHL ILLDMVDVGK PDLVLTLLCR WLHMVHLYSS RTSITSWIYS 660
661 IASKFYFIDS LNLETKDSVL ESNWERFISS TETYELDSIR DSLIDCLIRL LCYSSNNAID 720
721 PVSAPGNLDF DNSAELCSSL TRFTPKNIDF VLKSLIHIPI HSITKNQRTR LLNLLIIHEK 780
781 LLSDKDNSAH ISLRKIIYTL MKTPCSSGFY RDPKIIIDLM QFGKTDLSFH KVHVLIMRSW 840
841 IQHLDEGNKS EVILKLLQLI LADFRELPLE TITLIFNALV DSLPTSKHIK NSNDILSSVF 900
901 TINDMLIDEM TAYLSKKESL CSGTFSCLLA CCNSVLRTQN LTSESLNKLH ILKVGIEEKA 960
961 ISLKLNELPF LENWILFQCL LSFGGDDFLK VVAYMVYYRR TVGEMSFRVS DFELALSRLS1020
1021 TNDADYAILN ILNLLTSNEC DEFDYSVLMK VVGVFANYSD CFQRNRHLVH MVLLCCTHVF1080
1081 YNQTLKPATL IDSALDVFLC FVQKTSKFIS LDSFNTILTA LSTILNLKEL ECEPKDHANL1140
1141 ILKIIQILNG LLFNHRSYMS GRFHVFYSIL KSLLYSMVQH DSEMKKIKGI ALMNFYKPKK1200
1201 VPIHVVQSFC RLLTTWMNPN LMHDSNKKNS SLTDAERLFV KATTKHFVFL LTDFMSAQTV1260
1261 YQIDIQVKEL LTNCFYNVYE NLSNHEKQMA LVAMNAPSKS LYQKFVNDYL IFAKWNEA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
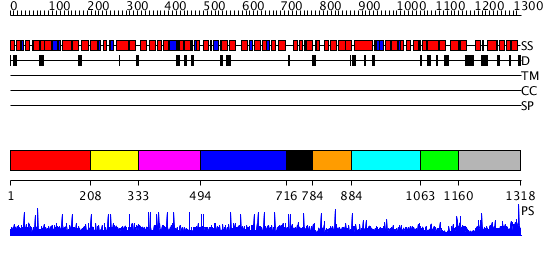
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [208..332] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [333..493] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [494..715] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [716..783] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [784..883] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [884..1062] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1063..1159] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1160..1318] | 1.009967 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)