













| General Information: |
|
| Name(s) found: |
POM1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1087 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGYLQSQKAV SLGDENTDAL FKLHTSNRKS ANMFGIKSEL LNPSELSAVG SYSNDICPNR 60
61 QSSSSTAADT SPSTNASNTN ISFPEQEHKD ELFMNVEPKG VGSSMDNHAI TIHHSTGNGL 120
121 LRSSFDHDYR QKNSPRNSIH RLSNISIGNN PIDFESSQQN NPSSLNTSSH HRTSSISNSK 180
181 SFGTSLSYYN RSSKPSDWNQ QNNGGHLSGV ISITQDVSSV PLQSSVFSSG NHAYHASMAP 240
241 KRSGSWRHTN FHSTSHPRAA SIGNKSGIPP VPTIPPNIGH STDHQHPKAN ISGSLTKSSS 300
301 ESKNLSTIQS PLKTSNSFFK ELSPHSQITL SNVKNNHSHV GSQTKSHSFA TPSVFDNNKP 360
361 VSSDNHNNTT TSSQVHPDSR NPDPKAAPKA VSQKTNVDGH RNHEAKHGNT VQNESKSQKS 420
421 SNKEGRSSRG GFFSRLSFSR SSSRMKKGSK AKHEDAPDVP AIPHAYIADS STKSSYRNGK 480
481 KTPTRTKSRM QQFINWFKPS KERSSNGNSD SASPPPVPRL SITRSQVSRE PEKPEEIPSV 540
541 PPLPSNFKDK GHVPQQRSVS YTPKRSSDTS ESLQPSLSFA SSNVLSEPFD RKVADLAMKA 600
601 INSKRINKLL DDAKVMQSLL DRACIITPVR NTEVQLINTA PLTEYEQDEI NNYDNIYFTG 660
661 LRNVDKRRSA DENTSSNFGF DDERGDYKVV LGDHIAYRYE VVDFLGKGSF GQVLRCIDYE 720
721 TGKLVALKII RNKKRFHMQA LVETKILQKI REWDPLDEYC MVQYTDHFYF RDHLCVATEL 780
781 LGKNLYELIK SNGFKGLPIV VIKSITRQLI QCLTLLNEKH VIHCDLKPEN ILLCHPFKSQ 840
841 VKVIDFGSSC FEGECVYTYI QSRFYRSPEV ILGMGYGTPI DVWSLGCIIA EMYTGFPLFP 900
901 GENEQEQLAC IMEIFGPPDH SLIDKCSRKK VFFDSSGKPR PFVSSKGVSR RPFSKSLHQV 960
961 LQCKDVSFLS FISDCLKWDP DERMTPQQAA QHDFLTGKQD VRRPNTAPAR QKFARPPNIE1020
1021 TAPIPRPLPN LPMEYNDHTL PSPKEPSNQA SNLVRSSDKF PNLLTNLDYS IISDNGFLRK1080
1081 PVEKSRP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
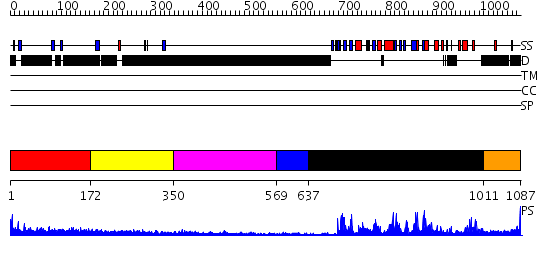
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.17 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | |
| 2 | View Details | [172..349] | 1.242996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [350..568] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [569..636] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [637..1010] | 69.30103 | Crystal structure of the phosphorylated CLK3 | |
| 6 | View Details | [1011..1087] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)