













| General Information: |
|
| Name(s) found: |
ILV2 /
YMR108W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA acetolactate synthase complex [IDA |
| Biological Process: |
branched chain family amino acid biosynthetic process
[IDA |
| Molecular Function: |
acetolactate synthase activity
[IDA FAD binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRQSTLKNF AIKRCFQHIA YRNTPAMRSV ALAQRFYSSS SRYYSASPLP ASKRPEPAPS 60
61 FNVDPLEQPA EPSKLAKKLR AEPDMDTSFV GLTGGQIFNE MMSRQNVDTV FGYPGGAILP 120
121 VYDAIHNSDK FNFVLPKHEQ GAGHMAEGYA RASGKPGVVL VTSGPGATNV VTPMADAFAD 180
181 GIPMVVFTGQ VPTSAIGTDA FQEADVVGIS RSCTKWNVMV KSVEELPLRI NEAFEIATSG 240
241 RPGPVLVDLP KDVTAAILRN PIPTKTTLPS NALNQLTSRA QDEFVMQSIN KAADLINLAK 300
301 KPVLYVGAGI LNHADGPRLL KELSDRAQIP VTTTLQGLGS FDQEDPKSLD MLGMHGCATA 360
361 NLAVQNADLI IAVGARFDDR VTGNISKFAP EARRAAAEGR GGIIHFEVSP KNINKVVQTQ 420
421 IAVEGDATTN LGKMMSKIFP VKERSEWFAQ INKWKKEYPY AYMEETPGSK IKPQTVIKKL 480
481 SKVANDTGRH VIVTTGVGQH QMWAAQHWTW RNPHTFITSG GLGTMGYGLP AAIGAQVAKP 540
541 ESLVIDIDGD ASFNMTLTEL SSAVQAGTPV KILILNNEEQ GMVTQWQSLF YEHRYSHTHQ 600
601 LNPDFIKLAE AMGLKGLRVK KQEELDAKLK EFVSTKGPVL LEVEVDKKVP VLPMVAGGSG 660
661 LDEFINFDPE VERQQTELRH KRTGGKH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
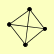
ILV2 ILV6 |
| View Details | Krogan NJ, et al. (2006) |
|
ILV2 PEX19 YDR053W YHR213W, YAR062W |
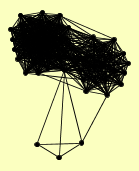
| View Details | Ho Y, et al. (2002) |
|
AAC3 AHA1 ATP3 AVO1 BEM2 CCT5 CLU1 DHH1 DPB2 ECM10 FUN12 GAL7 GCD11 GPH1 HOM3 HOR2 IDH1 ILV2 MET16 MKK2 MLH3 MLP2 NOG2 NPA3 OYE2 PDX1 PHO85 PIL1 PRB1 PTC3 PUF3 RAD52 RAD59 RHR2 RPN12 SEC27 SEC53 SIS1 SLX1 SRP54 STI1 TEF4 TEM1 TEP1 TFC7 UBA1 VMA8 YER077C YER138C YGR266W YKR051W YKU80 YLR271W YPR003C YPT31 |
| View Details | Qiu et al. (2008) |

|
ILV2 ILV6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..274] | 11000.0 | Acetohydroxyacid synthase catalytic subunit | |
| 2 | View Details | [275..460] [510..522] |
11000.0 | Acetohydroxyacid synthase catalytic subunit | |
| 3 | View Details | [461..509] [523..687] |
11000.0 | Acetohydroxyacid synthase catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)