













| General Information: |
|
| Name(s) found: |
ATG17 /
YLR423C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
autophagy
[IMP telomere maintenance [IMP |
| Molecular Function: |
kinase activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEADVTKFV NNARKTLTDA QLLCSSANLR IVDIKKKLSS WQLSISKLNF LIVGLRQQGK 60
61 FLYTILKEGI GTKLIQKQWN QAVLVVLVDE MKYWQYEITS KVQRLDGIVN ELSISEKDDT 120
121 DPSKLGDYIS RDNVNLLNDK LKEVPVIERQ IENIKLQYEN MVRKVNKELI DTKLTDVTQK 180
181 FQSKFGIDNL METNVAEQFS RELTDLEKDL AEIMNSLTQH FDKTLLLQDK KIDNDEREEL 240
241 FKVVQGDDKE LYNIFKTLHE VIDDVDKTIL NLGQFLQAKI KEKTELHSEV SEIINDFNRN 300
301 LEYLLIFKDI SNLIDSFKNS CTQDIQTTKE LCEFYDNFEE SYGNLVLEAK RRKDVANRMK 360
361 TILKDCEKQL QNLDAQDQEE RQNFIAENGT YLPETIWPGK IDDFSSLYTL NYNVKNP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ATG11 ATG17 ATG29 CIS1 |
| View Details | Krogan NJ, et al. (2006) |

|
ATG11 ATG17 ATG29 CIS1 THI11 YBR197C |
| View Details | Gavin AC, et al. (2006) |
|
ATG11 ATG17 ATG29 |
| View Details | Ho Y, et al. (2002) |

|
AAC1 AAC3 ADE5,7 AHA1 ARC1 ARO1 ARP2 ARP4 ASA1 ATG11 ATG12 ATG17 ATG29 ATP3 CAR2 CDC33 CIC1 CIS1 CPA2 CPR6 CYS4 FET4 FOL2 GCD11 GFA1 GRH1 GUS1 HAP2 HAP5 HHF1, HHF2 IPI3 IPP1 LOC1 MAM33 MTQ1 NAP1 NMD5 NOP16 NSA2 POL5 PPX1 PRB1 PSE1 PTC7 Q0032 REP1 REX2 RHR2 RPN10 RPN11 RPN3 RPN5 RPN6 RPN7 RPT1 RPT3 SAH1 SAP190 SEC18 SPE3 SSK2 TCB1 TIF1, TIF2 TIF6 TYR1 YHR020W YHR033W YNL208W YRA1 YRA2 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
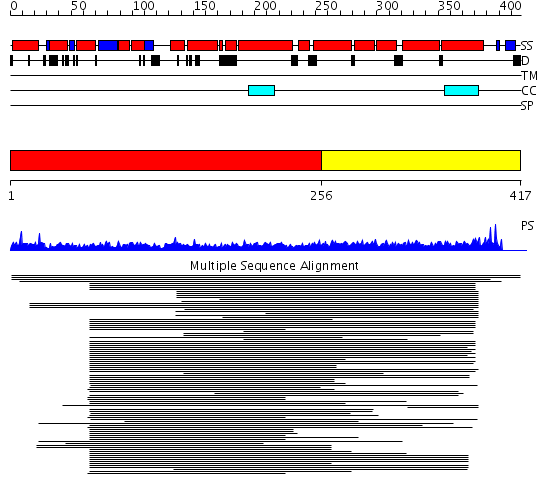
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..255] | 13.09691 | Kinesin | |
| 2 | View Details | [256..417] | 8.03 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)