













| General Information: |
|
| Name(s) found: |
TOP3 /
YLR234W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 656 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC RecQ helicase-Topo III complex [IDA |
| Biological Process: |
reciprocal meiotic recombination
[IGI telomere maintenance via telomerase [IMP regulation of DNA recombination [IMP |
| Molecular Function: |
DNA topoisomerase type I activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVLCVAEKN SIAKAVSQIL GGGRSTSRDS GYMYVKNYDF MFSGFPFARN GANCEVTMTS 60
61 VAGHLTGIDF SHDSHGWGKC AIQELFDAPL NEIMNNNQKK IASNIKREAR NADYLMIWTD 120
121 CDREGEYIGW EIWQEAKRGN RLIQNDQVYR AVFSHLERQH ILNAARNPSR LDMKSVHAVG 180
181 TRIEIDLRAG VTFTRLLTET LRNKLRNQAT MTKDGAKHRG GNKNDSQVVS YGTCQFPTLG 240
241 FVVDRFERIR NFVPEEFWYI QLVVENKDNG GTTTFQWDRG HLFDRLSVLT FYETCIETAG 300
301 NVAQVVDLKS KPTTKYRPLP LTTVELQKNC ARYLRLNAKQ SLDAAEKLYQ KGFISYPRTE 360
361 TDTFPHAMDL KSLVEKQAQL DQLAAGGRTA WASYAASLLQ PENTSNNNKF KFPRSGSHDD 420
421 KAHPPIHPIV SLGPEANVSP VERRVYEYVA RHFLACCSED AKGQSMTLVL DWAVERFSAS 480
481 GLVVLERNFL DVYPWARWET TKQLPRLEMN ALVDIAKAEM KAGTTAPPKP MTESELILLM 540
541 DTNGIGTDAT IAEHIDKIQV RNYVRSEKVG KETYLQPTTL GVSLVHGFEA IGLEDSFAKP 600
601 FQRREMEQDL KKICEGHASK TDVVKDIVEK YRKYWHKTNA CKNTLLQVYD RVKASM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Qiu et al. (2008) |
|
SGS1 TOP3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
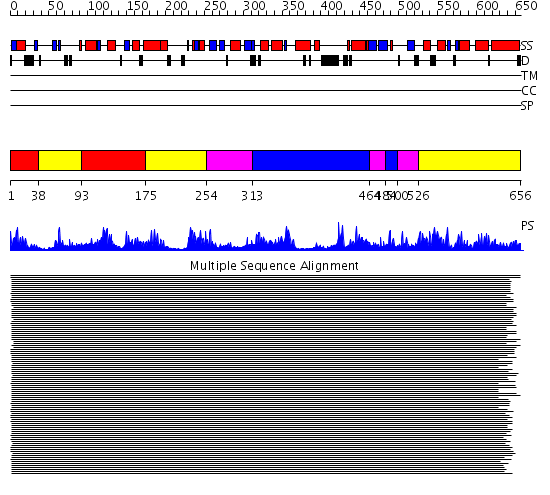
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..37] [93..174] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 2 | View Details | [38..92] [175..253] [526..656] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 3 | View Details | [254..312] [464..483] [500..525] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 4 | View Details | [313..463] [484..499] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)