













| General Information: |
|
| Name(s) found: |
ATP7 /
YKL016C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 174 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial proton-transporting ATP synthase, stator stalk
[IMP mitochondrion [IDA |
| Biological Process: |
protein complex assembly
[IMP ATP synthesis coupled proton transport [IMP |
| Molecular Function: |
hydrogen ion transporting ATP synthase activity, rotational mechanism
[IMP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
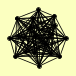
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ATP1 ATP14 ATP15 ATP16 ATP17 ATP18 ATP19 ATP2 ATP20 ATP3 ATP4 ATP5 ATP6 ATP7 ATP8 INH1 OLI1 TIM11 |
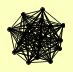
| View Details | Ho Y, et al. (2002) |
|
ADK1 ATP3 ATP5 ATP7 CDC14 DPM1 FUR1 GLC7 HEF3 HMS1 MCR1 SNF4 SPE3 SSZ1 TPS1 TSA2 VAS1 |
| View Details | Qiu et al. (2008) |

|
ATP1 ATP14 ATP15 ATP16 ATP17 ATP18 ATP19 ATP2 ATP20 ATP3 ATP4 ATP5 ATP6 ATP7 ATP8 COB COX1 COX12 COX13 COX2 COX3 COX4 COX5A COX5B COX6 COX7 COX8 COX9 INH1 MIR1 OLI1 QCR10 QCR7 QCR8 RIP1 STF1 STF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | De Craene, J., et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
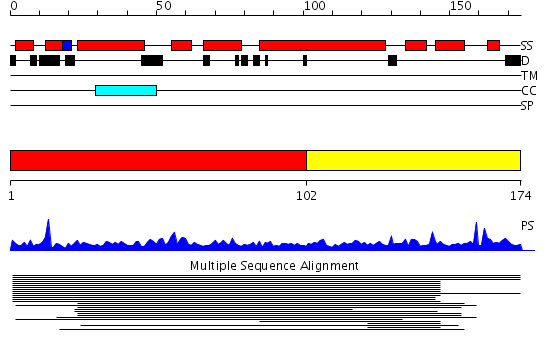
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 1.68 | Tropomyosin | |
| 2 | View Details | [102..174] | 1.68 | Tropomyosin |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)