













| General Information: |
|
| Name(s) found: |
SMC3 /
YJL074C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1230 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear cohesin complex [TAS |
| Biological Process: |
ascospore formation
[IMP mitotic sister chromatid cohesion [IMP synaptonemal complex assembly [IMP |
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYIKRVIIKG FKTYRNETII DNFSPHQNVI IGSNGSGKSN FFAAIRFVLS DDYSNLKREE 60
61 RQGLIHQGSG GSVMSASVEI VFHDPDHSMI LPSGVLSRGD DEVTIRRTVG LKKDDYQLND 120
121 RNVTKGDIVR MLETAGFSMN NPYNIVPQGK IVALTNAKDK ERLQLLEDVV GAKSFEVKLK 180
181 ASLKKMEETE QKKIQINKEM GELNSKLSEM EQERKELEKY NELERNRKIY QFTLYDRELN 240
241 EVINQMERLD GDYNNTVYSS EQYIQELDKR EDMIDQVSKK LSSIEASLKI KNATDLQQAK 300
301 LRESEISQKL TNVNVKIKDV QQQIESNEEQ RNLDSATLKE IKSIIEQRKQ KLSKILPRYQ 360
361 ELTKEEAMYK LQLASLQQKQ RDLILKKGEY ARFKSKDERD TWIHSEIEEL KSSIQNLNEL 420
421 ESQLQMDRTS LRKQYSAIDE EIEELIDSIN GPDTKGQLED FDSELIHLKQ KLSESLDTRK 480
481 ELWRKEQKLQ TVLETLLSDV NQNQRNVNET MSRSLANGII NVKEITEKLK ISPESVFGTL 540
541 GELIKVNDKY KTCAEVIGGN SLFHIVVDTE ETATLIMNEL YRMKGGRVTF IPLNRLSLDS 600
601 DVKFPSNTTT QIQFTPLIKK IKYEPRFEKA VKHVFGKTIV VKDLGQGLKL AKKHKLNAIT 660
661 LDGDRADKRG VLTGGYLDQH KRTRLESLKN LNESRSQHKK ILEELDFVRN ELNDIDTKID 720
721 QVNGNIRKVS NDRESVLTNI EVYRTSLNTK KNEKLILEES LNAIILKLEK LNTNRTFAQE 780
781 KLNTFENDLL QEFDSELSKE EKERLESLTK EISAAHNKLN ITSDALEGIT TTIDSLNAEL 840
841 ESKLIPQEND LESKMSEVGD AFIFGLQDEL KELQLEKESV EKQHENAVLE LGTVQREIES 900
901 LIAEETNNKK LLEKANNQQR LLLKKLDNFQ KSVEKTMIKK TTLVTRREEL QQRIREIGLL 960
961 PEDALVNDFS DITSDQLLQR LNDMNTEISG LKNVNKRAFE NFKKFNERRK DLAERASELD1020
1021 ESKDSIQDLI VKLKQQKVNA VDSTFQKVSE NFEAVFERLV PRGTAKLIIH RKNDNANDHD1080
1081 ESIDVDMDAE SNESQNGKDS EIMYTGVSIS VSFNSKQNEQ LHVEQLSGGQ KTVCAIALIL1140
1141 AIQMVDPASF YLFDEIDAAL DKQYRTAVAT LLKELSKNAQ FICTTFRTDM LQVADKFFRV1200
1201 KYENKISTVI EVNREEAIGF IRGSNKFAEV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
IRR1 MCD1 SMC1 SMC3 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
IRR1 MCD1 REC8 SCC2 SMC1 SMC3 |
| View Details | Krogan NJ, et al. (2006) |

|
IRR1 MCD1 SMC1 SMC3 |
| View Details | Gavin AC, et al. (2006) |

|
SMC1 SMC3 |
| View Details | Gavin AC, et al. (2002) |
|
ACC1 CCT8 CLU1 CPA2 FDH1 GCN1 GCN20 GFA1 GUS1 HHF1, HHF2 ILV1 IRR1 KAP123 LYS12 MCD1 MRPL3 NOP12 PSA1 RPA135 RPB2 RPN10 RPN11 RPN12 RPN8 RPT3 RPT5 RPT6 RVB2 SAM1 SCC4 SEC27 SMC1 SMC3 TCB1 TIF1, TIF2 TSA1 TUB3 URA7 VAS1 YHB1 YOL098C |
| View Details | Ho Y, et al. (2002) |
|
ASF1 CDC5 COP1 GFA1 GID8 GSY1 HTA2 IPP1 IRR1 IVY1 KAP95 MCD1 MDH1 NOP13 NUG1 PAA1 RAD53 RFC4 SMC1 SMC3 SPC72 SRP1 UBR1 URA7 UTP22 YGL081W |
| View Details | Qiu et al. (2008) |

|
REC8 SMC3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Huh WK, et al. (2003) |
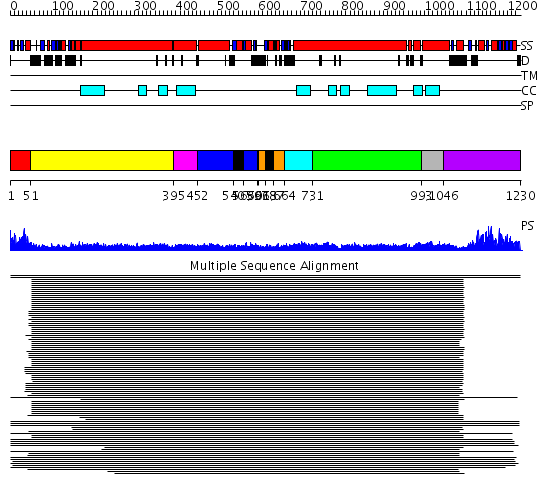
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..50] | 2.69897 | Myosin S1 fragment, N-terminal domain; Myosin S1, motor domain | |
| 2 | View Details | [51..394] | 22.522879 | Heavy meromyosin subfragment | |
| 3 | View Details | [395..451] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [452..539] [563..596] |
91.154902 | Smc hinge domain | |
| 5 | View Details | [540..562] [597..600] [618..636] |
91.154902 | Smc hinge domain | |
| 6 | View Details | [601..617] [637..663] |
91.154902 | Smc hinge domain | |
| 7 | View Details | [664..730] | 9.045757 | Tropomyosin | |
| 8 | View Details | [731..992] | 9.045757 | Tropomyosin | |
| 9 | View Details | [993..1045] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1046..1230] | 144.94696 | Smc head domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)