













| General Information: |
|
| Name(s) found: |
ERG11 /
YHR007C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 530 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
ergosterol biosynthetic process
[TAS]
|
| Molecular Function: |
sterol 14-demethylase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSATKSIVGE ALEYVNIGLS HFLALPLAQR ISLIIIIPFI YNIVWQLLYS LRKDRPPLVF 60
61 YWIPWVGSAV VYGMKPYEFF EECQKKYGDI FSFVLLGRVM TVYLGPKGHE FVFNAKLADV 120
121 SAEAAYAHLT TPVFGKGVIY DCPNSRLMEQ KKFVKGALTK EAFKSYVPLI AEEVYKYFRD 180
181 SKNFRLNERT TGTIDVMVTQ PEMTIFTASR SLLGKEMRAK LDTDFAYLYS DLDKGFTPIN 240
241 FVFPNLPLEH YRKRDHAQKA ISGTYMSLIK ERRKNNDIQD RDLIDSLMKN STYKDGVKMT 300
301 DQEIANLLIG VLMGGQHTSA ATSAWILLHL AERPDVQQEL YEEQMRVLDG GKKELTYDLL 360
361 QEMPLLNQTI KETLRMHHPL HSLFRKVMKD MHVPNTSYVI PAGYHVLVSP GYTHLRDEYF 420
421 PNAHQFNIHR WNKDSASSYS VGEEVDYGFG AISKGVSSPY LPFGGGRHRC IGEHFAYCQL 480
481 GVLMSIFIRT LKWHYPEGKT VPPPDFTSMV TLPTGPAKII WEKRNPEQKI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ALD6 CCT8 CYS4 ERG11 ERR3 HSP150 SLD2 TUB2 WAR1 |
| View Details | Qiu et al. (2008) |

|
ERG1 ERG11 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
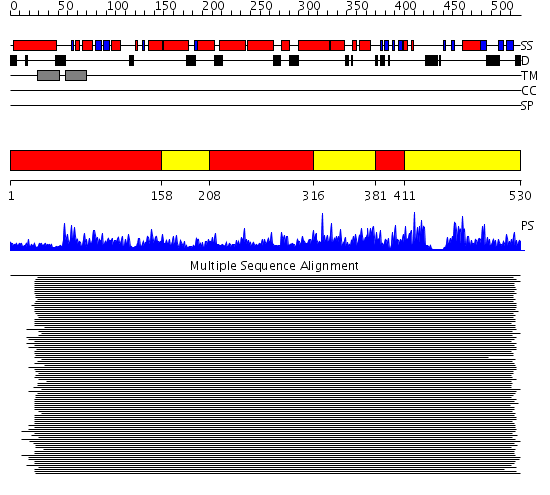
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] [208..315] [381..410] |
555.9794 | Cytochrome p450 14 alpha-sterol demethylase (cyp51) | |
| 2 | View Details | [158..207] [316..380] [411..530] |
555.9794 | Cytochrome p450 14 alpha-sterol demethylase (cyp51) |
Functions predicted (by domain):
| Source: Reynolds et al. 2008. Manuscript submitted |

|