













| General Information: |
|
| Name(s) found: |
NSR1 /
YGR159C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 414 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA nucleolus [TAS |
| Biological Process: |
telomere maintenance
[IMP ribosomal small subunit assembly [TAS rRNA processing [TAS |
| Molecular Function: |
RNA binding
[ISS single-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKTTKVKGN KKEVKASKQA KEEKAKAVSS SSSESSSSSS SSSESESESE SESESSSSSS 60
61 SSDSESSSSS SSDSESEAET KKEESKDSSS SSSDSSSDEE EEEEKEETKK EESKESSSSD 120
121 SSSSSSSDSE SEKEESNDKK RKSEDAEEEE DEESSNKKQK NEETEEPATI FVGRLSWSID 180
181 DEWLKKEFEH IGGVIGARVI YERGTDRSRG YGYVDFENKS YAEKAIQEMQ GKEIDGRPIN 240
241 CDMSTSKPAG NNDRAKKFGD TPSEPSDTLF LGNLSFNADR DAIFELFAKH GEVVSVRIPT 300
301 HPETEQPKGF GYVQFSNMED AKKALDALQG EYIDNRPVRL DFSSPRPNND GGRGGSRGFG 360
361 GRGGGRGGNR GFGGRGGARG GRGGFRPSGS GANTAPLGRS RNTASFAGSK KTFD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
NSR1 PRP43 RPL15A RPL4B RPS1B RPS4B, RPS4A UTP22 YGR054W |
| View Details | Hazbun TR, et al. (2003) |

|
GCD6 NOP9 NSR1 SNU13 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
DRS1 GAR1 IFH1 NOP1 NPL3 NSR1 POP1 POP2 RAT1 RNT1 RRP3 SBP1 SNM1 SRD1 UTP10 UTP11 UTP13 UTP4 UTP5 UTP6 UTP7 UTP8 UTP9 YME2 |
| View Details | Ho Y, et al. (2002) |
|
GLC7 HXT6 KOG1 NET1 NOP6 NSR1 PMA1 PMA2 PPZ2 REG1 RSE1 RVB1 SDS22 SNF4 YGR130C |
| View Details | Qiu et al. (2008) |

|
BRX1 CIC1 DBP3 DBP6 DBP7 DBP8 DHR2 DRS1 ECM16 ENP1 ERB1 FAL1 FOB1 GAR1 HCA4 IFH1 KRI1 KRR1 MAK11 MAK5 MRD1 MTR4 NAN1 NOP12 NOP13 NOP15 NOP2 NOP4 NOP7 NOP8 NOP9 NSR1 NUG1 PRP43 PUF6 PWP1 PWP2 PXR1 RNT1 RRB1 RRP1 RRP3 RRP5 RRP9 SOF1 SPB4 SSF1 TIF6 TRF5 UTP10 UTP11 UTP13 UTP15 UTP18 UTP4 UTP5 UTP6 UTP7 UTP8 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | control 2 | McCusker D, et al (2007) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
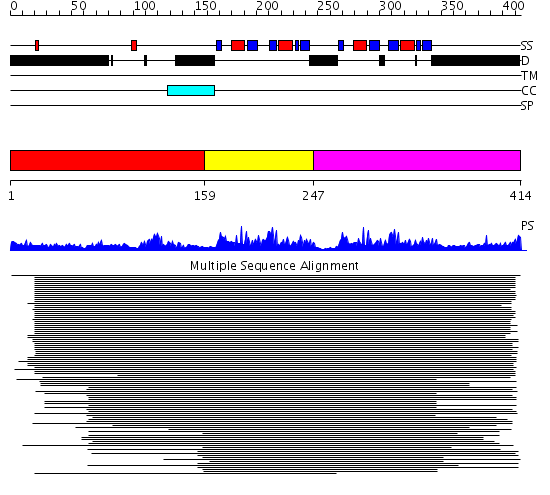
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 8.46 | No description for 1ldvA was found. | |
| 2 | View Details | [159..246] | 189.156545 | Poly(A)-binding protein | |
| 3 | View Details | [247..414] | 189.156545 | Poly(A)-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)