













| General Information: |
|
| Name(s) found: |
TFC4 /
YGR047C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1025 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor TFIIIC complex
[TAS |
| Biological Process: |
transcription initiation from RNA polymerase III promoter
[TAS |
| Molecular Function: |
RNA polymerase III transcription factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL 60
61 SEDEYNSERD SSLLAEFSDY GEISEDDEED FMNAIREASN FKVKKKKKND KGKSYGRQRK 120
121 ERVLDPEVAQ LLSQANEAFV RNDLQVAERL FNEVIKKDAR NFAAYETLGD IYQLQGRLND 180
181 CCNSWFLAAH LNASDWEFWK IVAILSADLD HVRQAIYCFS RVISLNPMEW ESIYRRSMLY 240
241 KKTGQLARAL DGFQRLYMYN PYDANILREL AILYVDYDRI EDSIELYMKV FNANVERREA 300
301 ILAALENALD SSDEESAAEG EDADEKEPLE QDEDRQMFPD INWKKIDAKY KCIPFDWSSL 360
361 NILAELFLKL AVSEVDGIKT IKKCARWIQR RESQTFWDHV PDDSEFDNRR FKNSTFDSLL 420
421 AAEKEKSYNI PIDIRVRLGL LRLNTDNLVE ALNHFQCLYD ETFSDVADLY FEAATALTRA 480
481 EKYKEAIDFF TPLLSLEEWR TTDVFKPLAR CYKEIESYET AKEFYELAIK SEPDDLDIRV 540
541 SLAEVYYRLN DPETFKHMLV DVVEMRKHQV DETLHRISNE KSSNDTSDIS SKPLLEDSKF 600
601 RTFRKKKRTP YDAERERIER ERRITAKVVD KYEKMKKFEL NSGLNEAKQA SIWINTVSEL 660
661 VDIFSSVKNF FMKSRSRKFV GILRRTKKFN TELDFQIERL SKLAEGDSVF EGPLMEERVT 720
721 LTSATELRGL SYEQWFELFM ELSLVIAKYQ SVEDGLSVVE TAQEVNVFFQ DPERVKMMKF 780
781 VKLAIVLQMD DEEELAENLR GLLNQFQFNR KVLQVFMYSL CRGPSSLNIL SSTIQQKFFL 840
841 RQLKAFDSCR YNTEVNGQAS ITNKEVYNPN KKSSPYLYYI YAVLLYSSRG FLSALQYLTR 900
901 LEEDIPDDPM VNLLMGLSHI HRAMQRLTAQ RHFQIFHGLR YLYRYHKIRK SLYTDLEKQE 960
961 ADYNLGRAFH LIGLVSIAIE YYNRVLENYD DGKLKKHAAY NSIIIYQQSG NVELADHLME1020
1021 KYLSI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
TFC1 TFC3 TFC4 TFC6 TFC7 TFC8 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
TFC1 TFC3 TFC4 TFC6 TFC7 |
| View Details | Krogan NJ, et al. (2006) |
|
TFC1 TFC3 TFC4 TFC6 TFC7 TFC8 |
| View Details | Gavin AC, et al. (2006) |
|
TFC1 TFC3 TFC4 TFC6 TFC7 TFC8 |
| View Details | Gavin AC, et al. (2002) |
|
ITC1 TFC1 TFC3 TFC4 TFC6 TFC7 TFC8 |
| View Details | Ho Y, et al. (2002) |
|
BUD14 CMP2 DOG1 GIS4 HSP104 KEL1 KEL2 KIN2 KRE6 POP2 TEF4 TFC4 UBA1 |
| View Details | Qiu et al. (2008) |
|
BDP1 BRF1 PZF1 SPT15 SSL1 TAF2 TFA1 TFB5 TFC1 TFC3 TFC4 TFC6 TFC7 TFC8 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
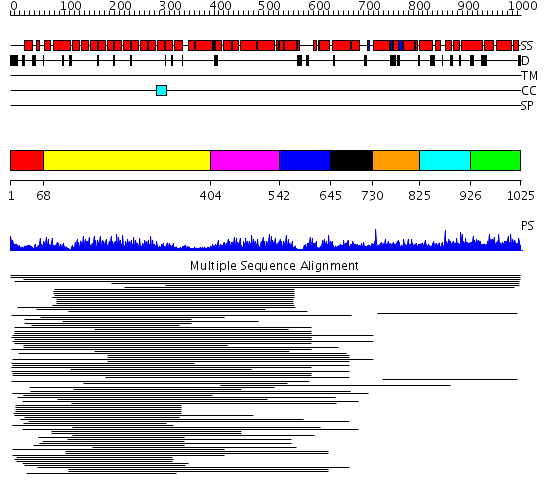
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 34.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [68..403] | 4.69897 | Clathrin heavy chain proximal leg segment | |
| 3 | View Details | [404..541] | 2.09691 | Vesicular transport protein sec17 | |
| 4 | View Details | [542..644] | 2.742997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [645..729] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [730..824] | 13.05 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 7 | View Details | [825..925] | 13.05 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 8 | View Details | [926..1025] | 13.05 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)