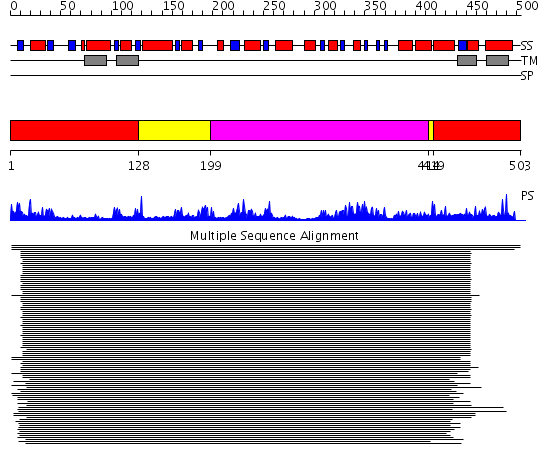
Description(s) found:
SHOW ONLY BEST
|
gi|6321373|ref|NP_011450.1| Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol;
[NCBI NR]
pir||S64069 ALG2 protein precursor - yeast (Saccharomyces cerevisiae)�
[NCBI NR]
gi|1703249|sp|P43636.2|ALG2_YEAST RecName: Full=Alpha-1,3-mannosyltransferase ALG2; AltName: Full=GDP-Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase; AltName: Full=Asparagine-linked glycosylation protein 2
[NCBI NR]
gi|1322572|emb|CAA96768.1| ALG2 [Saccharomyces cerevisiae]
[NCBI NR]
Alpha-1,3-mannosyltransferase ALG2 OS=Saccharomyces cerevisiae GN=ALG2 PE=1 SV=2
[Swiss-Prot]
ALG2 Mannosyltransferase
[mips]
Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol
[SGD]
ALG2, Chr VII from 379758-381269, reverse complement
[yeast_orfs2]
Chr VII from 379758-381269, reverse complement
[yeastorf3.fasta]
Chr VII from 379761-381272, reverse complement
[yeastorf4.fasta]
Chr VII from 381272-379761, reverse complement
[yeastorf5.fasta]
(P43636) Glycosyltransferase ALG2 (EC 2.4.1.-)
[4932.FASTACCERV.txt]
ALG2 SGDID:S0003033, Chr VII from 381272-379761, reverse complement, Verified ORF
[yeastflysequest.fasta]
ALG2 SGDID:S0003033, Chr VII from 381274-379763, reverse complement, Verified ORF
[10112005cervSGD.fasta]
Alg2p [Saccharomyces cerevisiae]�gi|1322572|emb|CAA96768.1| ALG2 [Saccharomyces cerevisiae]�gi|2131170|pir||S64069 ALG2 protein precursor - yeast (Saccharomyces cerevisiae)�gi|1703249|sp|P43636|ALG2_YEAST Alpha-1,3-mannosyltransferase ALG2 (GDP-Man:Man(1)
[nrdb_11152004_Chlamydomonas]
(P43636) Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP-Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase)
[cerv4932.FASTAC]
ALG2 SGDID:S000003033, Chr VII from 381274-379763, reverse complement, Verified ORF, "Presumed early mannosyltransferase involved in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lip
[SGD_S-cerevisiae_na_12-16-2005_con_reversed.fasta]
ALG2 SGDID:S000003033, Chr VII from 381274-379763, reverse complement, Verified ORF, "Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulati
[SGD_S-cerevisiae_na_08-19-2008_con_reversed.fasta]
ALG2 [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Alpha-1,3-mannosyltransferase ALG2 (GDP-Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase) (Asparagine-linked glycosylation protein 2)
[nr-271106-contam.fasta]
Presumed early mannosyltransferase involved in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol; Alg2p [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
ALG2 SGDID:S000003033, Chr VII from 381274-379763, reverse complement, Verified ORF, "Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol"
[yeast-bovrabprots.fasta]
ALG2 SGDID:S000003033, Chr VII from 381274-379763, reverse complement, Verified ORF, "Presumed early mannosyltransferase involved in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol"
[yeast-contam_custom.fasta]
[chemostatdb.fasta]
Alpha-1,3/1,6-mannosyltransferase ALG2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=ALG2 PE=1 SV=2
[UniProt_S_cerevisiae_20120516_05-16-2012_reversed.fasta]
ALG2 SGDID:S000003033, Chr VII from 381271-379760, Genome Release 64-1-1, reverse complement, Verified ORF, "Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol"
[yeast-030211-contam.fasta]
ALG2 SGDID:S000003033, Chr VII from 381271-379760, Genome Release 64-2-1, reverse complement, Verified ORF, "Mannosyltransferase in the N-linked glycosylation pathway; catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol"
[20141208_orf_trans.fasta]
YGL065C SGDID:S000003033, Chr VII from 381271-379760, Genome Release 64-1-1, reverse complement, Verified ORF, "Mannosyltransferase that catalyzes two consecutive steps in the N-linked glycosylation pathway; alg2 mutants exhibit temperature-sensitive growth and abnormal accumulation of the lipid-linked oligosaccharide Man2GlcNAc2-PP-Dol"
[2016-01-SGD_Yeast_Contam_Riffle_20140507-Ska-Hec-110-tubulin.fasta]
|