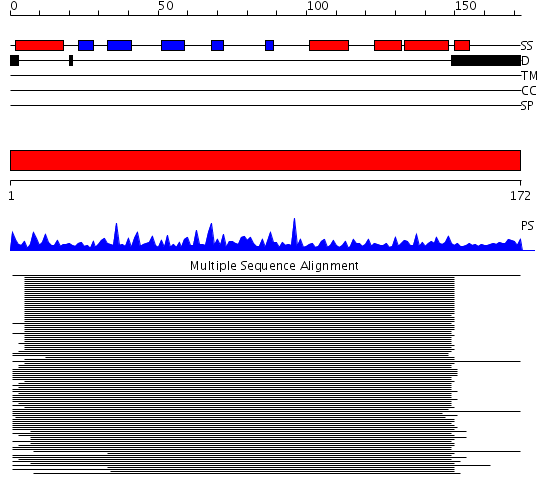
Description(s) found:
SHOW ONLY BEST
|
pir||A21906 ubiquitin-conjugating enzyme RAD6 - yeast (Saccharomyces cerevisiae)�
[NCBI NR]
gi|6321380|ref|NP_011457.1| Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (with Rad18p), sporulation, telomere silencing, and ubiquitin-mediated N-end rule protein degradation (with Ubr1p); Rad6p [Saccharomyces cerevisiae]
[NCBI NR]
gi|172349|gb|AAA34952.1| RAD6 protein
[NCBI NR]
gi|136641|sp|P06104.1|UBC2_YEAST RecName: Full=Ubiquitin-conjugating enzyme E2 2; AltName: Full=Ubiquitin-protein ligase UBC2; AltName: Full=Ubiquitin carrier protein UBC2; AltName: Full=Radiation sensitivity protein 6
[NCBI NR]
gi|1322558|emb|CAA96761.1| RAD6 [Saccharomyces cerevisiae]
[NCBI NR]
Ubiquitin-conjugating enzyme E2 2 OS=Saccharomyces cerevisiae GN=RAD6 PE=1 SV=1
[Swiss-Prot]
RAD6 E2 ubiquitin-conjugating enzyme
[mips]
Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (as a heterodimer with Rad18p), DSBR and checkpoint control (as a heterodimer with Bre1p), ubiquitin-mediated N-end rule protein degradation (as a heterodimer with Ubr1p
[SGD]
RAD6, Chr VII from 393986-394504
[yeast_orfs2]
Chr VII from 393986-394504
[yeastorf3.fasta]
Chr VII from 393990-394508
[yeastorf4.fasta]
(P06104) Ubiquitin-conjugating enzyme E2-20 kDa (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein)
[4932.FASTACCERV.txt]
RAD6 SGDID:S0003026, Chr VII from 393990-394508, Verified ORF
[yeastflysequest.fasta]
RAD6 SGDID:S0003026, Chr VII from 393992-394510, Verified ORF
[10112005cervSGD.fasta]
Rad6p [Saccharomyces cerevisiae]�gi|1322558|emb|CAA96761.1| RAD6 [Saccharomyces cerevisiae]�gi|83451|pir||A21906 ubiquitin-conjugating enzyme RAD6 - yeast (Saccharomyces cerevisiae)�gi|136641|sp|P06104|UBC2_YEAST Ubiquitin-conjugating enzyme E2-20 kDa (Ub
[nrdb_11152004_Chlamydomonas]
RAD6 SGDID:S000003026, Chr VII from 393992-394510, Verified ORF, "Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (with Rad18p), sporulation, telomere silencing, and ubiquitin-mediated N-end rule protein degradation (with Ubr1p)"
[SGD_S-cerevisiae_na_12-16-2005_con_reversed.fasta]
Ubiquitin-conjugating enzyme E2 2 (Ubiquitin-protein ligase UBC2) (Ubiquitin carrier protein UBC2) (Radiation sensitivity protein 6)
[nr-271106-contam.fasta]
RAD6 protein
[nr-271106-contam.fasta]
RAD6 [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (with Rad18p), sporulation, telomere silencing, and ubiquitin-mediated N-end rule protein degradation (with Ubr1p); Rad6p [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
RAD6 SGDID:S000003026, Chr VII from 393992-394510, Verified ORF, "Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (as a heterodimer with Rad18p), DSBR and checkpoint control (as a heterodimer with Bre1p), ubiquitin-mediated N-end rul
[SGD_S-cerevisiae_na_01-05-2010_reversed.fasta]
[chemostatdb.fasta]
RAD6 SGDID:S000003026, Chr VII from 393992-394510, Verified ORF, "Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (as a heterodimer with Rad18p), DSBR and checkpoint control (as a heterodimer with Bre1p), ubiquitin-mediated N-end rule protein degradation (as a heterodimer with Ubr1p"
[SGDyeast.20101221.fasta]
Ubiquitin-conjugating enzyme E2 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=RAD6 PE=1 SV=1
[UniProt_S_cerevisiae_20120516_05-16-2012_reversed.fasta]
RAD6 SGDID:S000003026, Chr VII from 393986-394504, Genome Release 64-1-1, Verified ORF, "Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (as a heterodimer with Rad18p), DSBR and checkpoint control (as a heterodimer with Bre1p), ubiquitin-mediated N-end rule protein degradation (as a heterodimer with Ubr1p"
[yeast-030211-contam.fasta]
RAD6 SGDID:S000003026, Chr VII from 393986-394504, Genome Release 64-2-1, Verified ORF, "Ubiquitin-conjugating enzyme (E2); involved in postreplication repair as a heterodimer with Rad18p, DSBR and checkpoint control as a heterodimer with Bre1p, ubiquitin-mediated N-end rule protein degradation as a heterodimer with Ubr1p, as well as endoplasmic reticulum-associated protein degradation (ERAD) with Ubr1p in the absence of canonical ER membrane ligases"
[20141208_orf_trans.fasta]
YGL058W SGDID:S000003026, Chr VII from 393986-394504, Genome Release 64-1-1, Verified ORF, "Ubiquitin-conjugating enzyme (E2), involved in postreplication repair (as a heterodimer with Rad18p), DSBR and checkpoint control (as a heterodimer with Bre1p), ubiquitin-mediated N-end rule protein degradation (as a heterodimer with Ubr1p"
[2016-01-SGD_Yeast_Contam_Riffle_20140507-Ska-Hec-110-tubulin.fasta]
|