













| General Information: |
|
| Name(s) found: |
UBP6 /
YFR010W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 499 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome regulatory particle
[IPI |
| Biological Process: |
protein deubiquitination
[TAS |
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGETFEFNI RHSGKVYPIT LSTDATSADL KSKAEELTQV PSARQKYMVK GGLSGEESIK 60
61 IYPLIKPGST VMLLGTPDAN LISKPAKKNN FIEDLAPEQQ VQQFAQLPVG FKNMGNTCYL 120
121 NATLQALYRV NDLRDMILNY NPSQGVSNSG AQDEEIHKQI VIEMKRCFEN LQNKSFKSVL 180
181 PIVLLNTLRK CYPQFAERDS QGGFYKQQDA EELFTQLFHS MSIVFGDKFS EDFRIQFKTT 240
241 IKDTANDNDI TVKENESDSK LQCHISGTTN FMRNGLLEGL NEKIEKRSDL TGANSIYSVE 300
301 KKISRLPKFL TVQYVRFFWK RSTNKKSKIL RKVVFPFQLD VADMLTPEYA AEKVKVRDEL 360
361 RKVEKEKNEK EREIKRRKFD PSSSENVMTP REQYETQVAL NESEKDQWLE EYKKHFPPNL 420
421 EKGENPSCVY NLIGVITHQG ANSESGHYQA FIRDELDENK WYKFNDDKVS VVEKEKIESL 480
481 AGGGESDSAL ILMYKGFGL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ECM29 GSG1 NAS6 OSH7 POL4 RPN1 RPN10 RPN11 RPN12 RPN13 RPN2 RPN3 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 SEC18 SEM1 SLX5 SPG5 SWA2 UBP6 URA3 VPS45 |
| View Details | Krogan NJ, et al. (2006) |

|
CSI1 CSN9 ERV46 FPR2 HUL5 IQG1 MOT1 MPT5 NAS6 NMD3 NPT1 PCI8 PEP1 PST1 PUS9 RPN1 RPN10 RPN11 RPN12 RPN13 RPN14 RPN2 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RRI1 RRI2 SEC18 SEM1 SKP1 SPG5 SPO77 SQT1 SWA2 TCB2 TIP41 TOD6 TOR2 TSR4 UBP6 YEN1 YKL027W YMR258C YNL022C YPR003C YPT52 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC6 ECM29 GFA1 MLH2 NAS6 PGK1 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP3 RGR1 RPN10 RPN11 RPN12 RPN13 RPN3 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 SCL1 UBP6 YFL006W |
| View Details | Qiu et al. (2008) |
|
DOA4 RPN12 RPN13 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 UBP6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
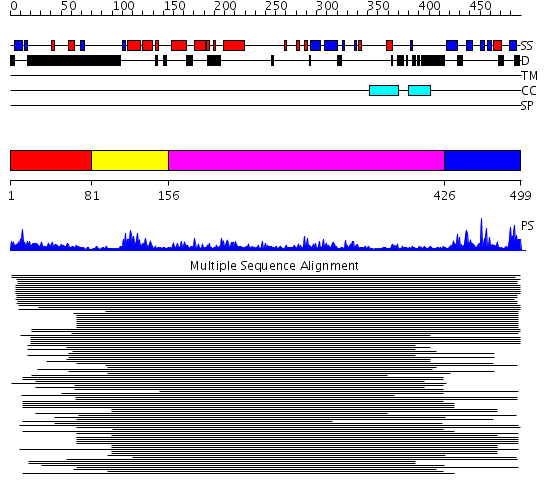
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 8.39794 | Ubiquitin | |
| 2 | View Details | [81..155] | 14.031517 | No description for PF00442 was found. No confident structure predictions are available. | |
| 3 | View Details | [156..425] | 6.241996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [426..499] | 29.619789 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)