













| General Information: |
|
| Name(s) found: |
ATG9 /
YDL149W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 997 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA cytoplasm [IDA mitochondrion [IDA integral to membrane [IDA |
| Biological Process: |
autophagic vacuole assembly
[IMP autophagy [IMP protein targeting to vacuole [IMP |
| Molecular Function: |
identical protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERDEYQLPN SHGKNTFLSR IFGLQSDEVN PSLNSQEMSN FPLPDIERGS SLLHSTNDSR 60
61 EDVDENDLRV PESDQGTSTE EEDEVDEEQV QAYAPQISDG LDGDHQLNSV TSKENVLETE 120
121 KSNLERLVEG STDDSVPKVG QLSSEEEEDN EFINNDGFDD DTPLFQKSKI HEFSSKKSNT 180
181 IEDGKRPLFF RHILQNNRPQ RDTQKLFTSS NAIHHDKDKS ANNGPRNING NQKHGTKYFG 240
241 SATQPRFTGS PLNNTNRFTK LFPLRKPNLL SNISVLNNTP EDRINTLSVK ERALWKWANV 300
301 ENLDIFLQDV YNYYLGNGFY CIILEKILNI CTLLFVVFVS TYMGHCVDYS KLPTSHRVSD 360
361 IIIDKCYSNS ITGFTKFFLW MFYFFVILKI VQLYFDVQKL SELQNFYKYL LNISDDELQT 420
421 LPWQNVIQQL MYLKDQNAMT ANVVEVKAKN RIDAHDVANR IMRRENYLIA LYNSDILNLS 480
481 LPIPLFRTNV LTKTLEWNIN LCVMGFVFNE SGFIKQSILK PSQREFTREE LQKRFMLAGF 540
541 LNIILAPFLV TYFVLLYFFR YFNEYKTSPG SIGARQYTPI AEWKFREYNE LYHIFKKRIS 600
601 LSTTLANKYV DQFPKEKTNL FLKFVSFICG SFVAILAFLT VFDPENFLNF EITSDRSVIF 660
661 YITILGAIWS VSRNTITQEY HVFDPEETLK ELYEYTHYLP KEWEGRYHKE EIKLEFCKLY 720
721 NLRIVILLRE LTSLMITPFV LWFSLPSSAG RIVDFFRENS EYVDGLGYVC KYAMFNMKNI 780
781 DGEDTHSMDE DSLTKKIAVN GSHTLNSKRR SKFTAEDHSD KDLANNKMLQ SYVYFMDDYS 840
841 NSENLTGKYQ LPAKKGYPNN EGDSFLNNKY SWRKQFQPGQ KPELFRIGKH ALGPGHNISP 900
901 AIYSTRNPGK NWDNNNNGDD IKNGTNNATA KNDDNNGNND HEYVLTESFL DSGAFPNHDV 960
961 IDHNKMLNSN YNGNGILNKG GVLGLVKEYY KKSDVGR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
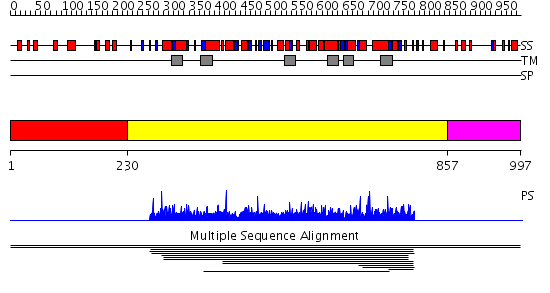
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..229] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [230..856] | 3.011 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [857..997] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|