













| General Information: |
|
| Name(s) found: |
UBX3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 410 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDREDILKEF CNRNNIDVSQ GRFFLESTNW NYELATALLH EVIPPEEDHG LQPSSDVSKV 60
61 PEVTGSSSGI SGGDQQPPRP LQRQQNTQGQ GMKSGTASKK FATLRDLEGN DESAEEKSHL 120
121 FTGGEKSGLS VEDGDPDPKK QLVRDILEKA RQHTISPLDE QDSGPSSLAS SWASVGQRLG 180
181 TENEASGSTT PVTQSGPPRE NPPTESQPEK PLRRTLYFWR NGFSVDDGPI YTYDDPANQE 240
241 MLRYINSGRA PLHLLGVSMN QPIDVVVQHR MDEDYVAPFK PFSGKGQRLG STYMQPRMSQ 300
301 MPGGLYTDTS TSSSVPINVK PNSTTPHASL QIDENKPTTR IQVRLSNGGR TVLTVNLSHT 360
361 LHDIYEAVRA VSPGNFILSV PFPAKTLEDD PSVTVEAASL KNASLVQKSL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
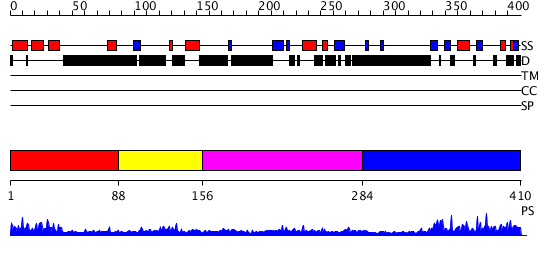
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 9.053997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [88..155] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [156..283] | 11.522879 | Solution structures of the p47 SEP domain | |
| 4 | View Details | [284..410] | 23.154902 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)