













| General Information: |
|
| Name(s) found: |
gi|61362057
[NCBI NR]
gi|168275772 [NCBI NR] gi|124000679 [NCBI NR] gi|123982872 [NCBI NR] gi|109451642 [NCBI NR] gi|109451064 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | synthetic construct |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSHLYRDG EGRIDDDDDE RENFEITDWD LQNEFNPNRQ RHWQTKEEAT YGVWAERDSD 60
61 DERPSFGGKR ARDYSAPVNF ISAGLKKGAA EEAELEDSDD EEKPVKQDDF PKDFGPRKLK 120
121 TGGNFKPSQK GFAGGTKSFM DFGSWERHTK GIGQKLLQKM GYVPGRGLGK NAQGIINPIE 180
181 AKQRKGKGAV GAYGSERTTQ SMQDFPVVDS EEEAEEEFQK ELSQWRKDPS GSKKKPKYSY 240
241 KTVEELKAKG RISKKLTAPQ KELSQVKVID MTGREQKVYY SYSQISHKHN VPDDGLPLQS 300
301 QQLPQSGKEA KAPGFALPEL EHNLQLLIDL TEQEIIQNDR QLQYERDMVV NLFHELEKMT 360
361 EVLDHEERVI SNLSKVLEMV EECERRMQPD CSNPLTLDEC ARIFETLQDK YYEEYRMSDR 420
421 VDLAVAIVYP LMKEYFKEWD PLKDCTYGTE IISKWKSLLE NDQLLSHGGQ DLSADAFHRL 480
481 IWEVWMPFVR NIVTQWQPRN CDPMVDFLDS WVHIIPVWIL DNILDQLIFP KLQKEVENWN 540
541 PLTDTVPIHS WIHPWLPLMQ ARLEPLYSPI RSKLSSALQK WHPSDSSAKL ILQPWKDVFT 600
601 PGSWEAFMVK NIVPKLGMCL GELVINPHQQ HMDAFYWVID WEGMISVSSL VGLLEKHFFP 660
661 KWLQVLCSWL SNSPNYEEIT KWYLGWKSMF SDQVLAHPSV KDKFNEALDI MNRAVSSNVG 720
721 AYMQPGAREN IAYLTHTERR KDFQYEAMQE RREAENMAQR GIGVAASSVP MNFKDLIETK 780
781 AEEHNIVFMP VIGKRHEGKQ LYTFGRIVIY IDRGVVFVQG EKTWVPTSLQ SLIDMAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
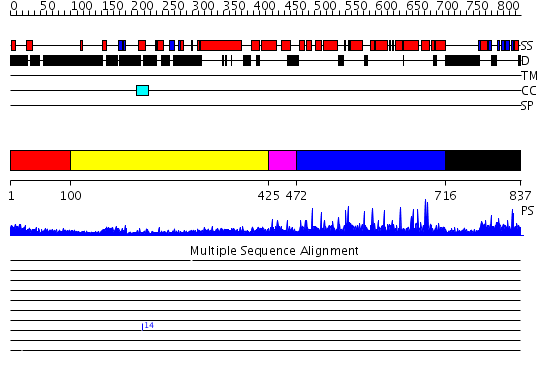
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..424] | 11.09691 | Heavy meromyosin subfragment | |
| 3 | View Details | [425..471] | 1.140995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [472..715] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [716..837] | 1.022987 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)