













| General Information: |
|
| Name(s) found: |
MUR3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[ISS]
integral to Golgi membrane [IDA] Golgi apparatus [IDA] |
| Biological Process: |
xyloglucan biosynthetic process
[IMP]
unidimensional cell growth [IMP] salicylic acid mediated signaling pathway [IMP] fucose biosynthetic process [IMP] endomembrane system organization [IMP] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[IDA]
catalytic activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFPRVSMRRR SAEVSPTEPM EKGNGKNQTN RICLLVALSL FFWALLLYFH FVVLGTSNID 60
61 KQLQLQPSYA QSQPSSVSLR VDKFPIEPHA APSKPPPKEP LVTIDKPILP PAPVANSSST 120
121 FKPPRIVESG KKQEFSFIRA LKTVDNKSDP CGGKYIYVHN LPSKFNEDML RDCKKLSLWT 180
181 NMCKFTTNAG LGPPLENVEG VFSDEGWYAT NQFAVDVIFS NRMKQYKCLT NDSSLAAAIF 240
241 VPFYAGFDIA RYLWGYNISR RDAASLELVD WLMKRPEWDI MRGKDHFLVA GRITWDFRRL 300
301 SEEETDWGNK LLFLPAAKNM SMLVVESSPW NANDFGIPYP TYFHPAKDSE VFEWQDRMRN 360
361 LERKWLFSFA GAPRPDNPKS IRGQIIDQCR NSNVGKLLEC DFGESKCHAP SSIMQMFQSS 420
421 LFCLQPQGDS YTRRSAFDSM LAGCIPVFFH PGSAYTQYTW HLPKNYTTYS VFIPEDDVRK 480
481 RNISIEERLL QIPAKQVKIM RENVINLIPR LIYADPRSEL ETQKDAFDVS VQAVIDKVTR 540
541 LRKNMIEGRT EYDYFVEENS WKYALLEEGQ REAGGHVWDP FFSKPKPGED GSSDGNGGTT 600
601 ISADAAKNSW KSEQRDKTQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
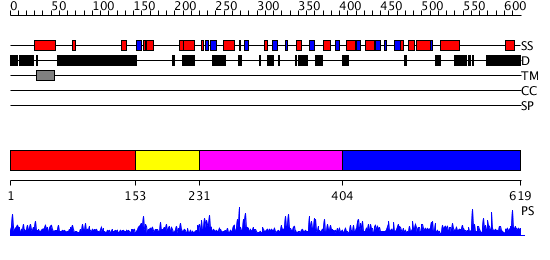
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [153..230] | 1.123987 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [231..403] | 1.133991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [404..619] | 5.142989 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|