













| General Information: |
|
| Name(s) found: |
gi|24372539
[NCBI NR]
gi|24346551 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 574 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA recombination
[ISS |
| Molecular Function: |
single-stranded DNA specific exodeoxyribonuclease activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIHKIIRRPT VDDSHLPTHL PPLLRQLYAR RGVLPSECEL VLKGLLRPDT MKGLDLAAKL 60
61 IADAMQLDKS ILIVGDFDAD GATSTSVCIL ALRMMGASKV DYLIPNRFDY GYGLSPEIVA 120
121 VAASKQAQLL ITVDNGISSI EGVAAAKAFG MQVVITDHHL PGHELPKADA IVNPNQPGCQ 180
181 FASKSIAGVG VAFYLMTALR AELRARNWYQ ARGIAEPNLG TLLDIVALGT VADVVSLDTN 240
241 NRILVEAGLQ RVRSGRCRVG ITALLEVAKR NPARIVASDF GFAVGPRLNA AGRLDEMALG 300
301 VETLLCEDMM YARRMAAELD SLNQERRELE VGMQQEALKS LEYLKLNEEQ LPWGIALFQE 360
361 DWHQGVIGIL ASRIKDKYHR PVIAFADAGN GEIKGSARSI KGLHMRDLLE LVNSRHPGLI 420
421 SKFGGHAMAA GLTLKSGGFA TFAKAYDDAV REQLKPEQLT GELWSDGELT PTELTLEIAQ 480
481 LLRNAGPWGQ SFEEPLFDGY FKIIQQRIVG DRHLKLVLET QCGTVMLDAI AFNVALHTWP 540
541 DATIQHARIV YKLDVNEYRG NFTLQLMVEQ IEPM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
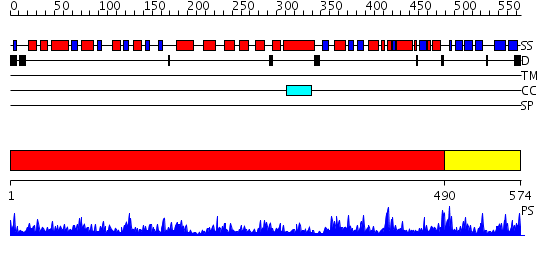
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..489] | 95.0 | Exonuclease RecJ | |
| 2 | View Details | [490..574] | 1.216979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)