













| General Information: |
|
| Name(s) found: |
gi|51854375
[NCBI NR]
gi|215701015 [NCBI NR] gi|115464181 [NCBI NR] gi|113579241 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 644 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADGASSSGA ASASGPGGGG GGESVVIDYG RRRTACGYCR STGQTSISHG LWANSLRADD 60
61 YQALLDRGWR RSGCFLYKPE MERTCCPQYT IRLKANDFIC SKEQGRVLKK MQRFLDGELD 120
121 PQVGSPQNKT SPTKHSLAEP MNMPVSKISK TLTNDFQAAK CPNLFEEDEF ISCLSSKINE 180
181 AVGMSFQVGT LGSDVQLPKA VVKTVKPQLK KKVGGASQDK KVGEAVQDLL YTCNISFQIV 240
241 AEIRRALPKE KDANHNKVVA DISPNSIAEK LAMTMECHGD IAGLAVKACN GHLNFYSVTN 300
301 QTKQNKTSII VSTHAPDKSS SSKQSSVNKN TVRVPQKRRK LEIKMRRSHF DPEEFALYQR 360
361 YQTKVHKEKK VSESSYKRFL VDTPIVFVPP RSGDNTVPPC GFGSFHQQYR IDGKLVAVGV 420
421 VDILPKCLSS KYLFWDPDFA FLSLGKYTAL KEIDWVKTTQ EHCPNLQYYY LGYYIHSCNK 480
481 MRYKAAYRPS ELLCPVRFEW VCYDSAKRLL DKSLYSVLSD FAQIQDEMPQ PQNSHLDTEL 540
541 SKNDNCESPI DEDDEDLSYD DSDMMVDEEM VRSESNTDVM EDCSSIIDFE NVMMDLNGSR 600
601 VKYKDLLGVV GRIERRHLEQ LERQLSKYVK VVGKELSDRM VYSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
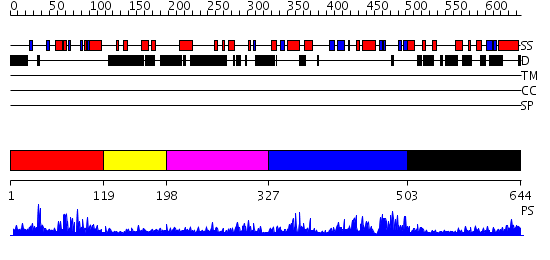
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 37.60206 | No description for PF04376.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [119..197] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [198..326] | 1.124989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [327..502] | 90.39794 | No description for PF04377.6 was found. No confident structure predictions are available. | |
| 5 | View Details | [503..644] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)