













| General Information: |
|
| Name(s) found: |
NILP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 326 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nitrogen compound metabolic process
[ISS]
putrescine biosynthetic process [TAS] |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
[ISS]
N-carbamoylputrescine amidase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METEGRRREV VVSSLQFACS DDISTNVAAA ERFVSLSSSL PLSNYQSLPS SSSFKFPYAL 60
61 VREAHAKGAN IILIQELFEG YYFCQAQRED FFKRAKPYKN HPTIARMQKL AKELGVVIPV 120
121 SFFEEANTAH YNSIAIIDAD GTDLGIYRKS HIPDGPGYQE KFYFNPGDTG FKVFQTKFAK 180
181 IGVAICWDQW FPEAARAMVL QGAEILFYPT AIGSEPQDQG LDSRDHWRRV MQGHAGANVV 240
241 PLVASNRIGK EIIETEHGPS QITFYGTSFI AGPTGEIVAE ADDKSEAVLV AQFDLDMIKS 300
301 KRQSWGVFRD RRPDLYKVLL TMDGNL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
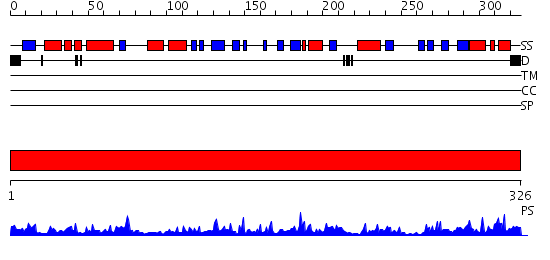
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..326] | 59.39794 | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)