













| General Information: |
|
| Name(s) found: |
ADO2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 611 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Cajal body
[IDA]
cytosol [IDA] |
| Biological Process: |
circadian rhythm
[ISS]
regulation of circadian rhythm [IMP] ubiquitin-dependent protein catabolic process [TAS] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNQMEWDSD SDLSGGDEVA EDGWFGGDNG AIPFPVGSLP GTAPCGFVVS DALEPDNPII 60
61 YVNTVFEIVT GYRAEEVIGR NCRFLQCRGP FTKRRHPMVD STIVAKMRQC LENGIEFQGE 120
121 LLNFRKDGSP LMNKLRLVPI REEDEITHFI GVLLFTDAKI DLGPSPDLSA KEIPRISRSF 180
181 TSALPIGERN VSRGLCGIFE LSDEVIAIKI LSQLTPGDIA SVGCVCRRLN ELTKNDDVWR 240
241 MVCQNTWGTE ATRVLESVPG AKRIGWVRLA REFTTHEATA WRKFSVGGTV EPSRCNFSAC 300
301 AVGNRIVIFG GEGVNMQPMN DTFVLDLGSS SPEWKSVLVS SPPPGRWGHT LSCVNGSRLV 360
361 VFGGYGSHGL LNDVFLLDLD ADPPSWREVS GLAPPIPRSW HSSCTLDGTK LIVSGGCADS 420
421 GALLSDTFLL DLSMDIPAWR EIPVPWTPPS RLGHTLTVYG DRKILMFGGL AKNGTLRFRS 480
481 NDVYTMDLSE DEPSWRPVIG YGSSLPGGMA APPPRLDHVA ISLPGGRILI FGGSVAGLDS 540
541 ASQLYLLDPN EEKPAWRILN VQGGPPRFAW GHTTCVVGGT RLVVLGGQTG EEWMLNEAHE 600
601 LLLATSTTAS T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
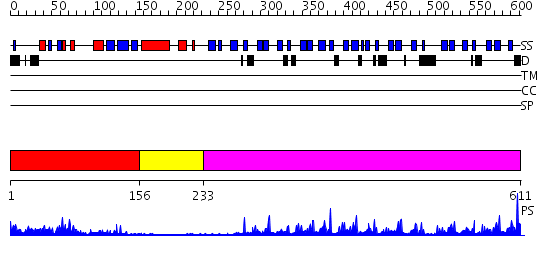
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 15.0 | No description for 2pr5A was found. | |
| 2 | View Details | [156..232] | 3.06 | No description for 2e31A was found. | |
| 3 | View Details | [233..611] | 39.69897 | Crystal structure of the Kelch domain of human Keap1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)