













| General Information: |
|
| Name(s) found: |
CYP37_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 466 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
thylakoid lumen [IDA] chloroplast thylakoid lumen [IDA][ISS] chloroplast thylakoid membrane [IDA] thylakoid [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASPLSSSTV VSHRLFFLHP SPLNRKFLFV KPKLPFNRTN SGDFRMRLHS TSSKTGTKEL 60
61 IHSCNSSIDS KLNTFEAGSK NLEKLVATIL IFVQVWSPLP LFGLDSAYIS PAEAVLYSPD 120
121 TKVPRTGELA LRRAIPANPS MKIIQASLED ISYLLRIPQR KPYGTMESNV KKALKVAIDD 180
181 KDKILASIPV DLKDKGSELY TTLIDGKGGL QALITSIKKQ DPDKVSLGLA ASLDTVADLE 240
241 LLQASGLSFL LPQQYLNYPR LAGRGTVEIT IEKADGSTFS AEAGGDQRKS ATVQIVIDGY 300
301 SAPLTAGNFA KLVTSGAYDG AKLNTVNQAV ITEDGSGKVE SVSVPLEVMP SGQFEPLYRT 360
361 PLSVQDGELP VLPLSVYGAV AMAHSENSEE YSSPYQFFFY LYDKRNSGLG GLSFDEGQFS 420
421 VFGYTIAGKD ILGQIKTGDI IKSAKLIEGQ DRLSLPVQNN NINEST |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
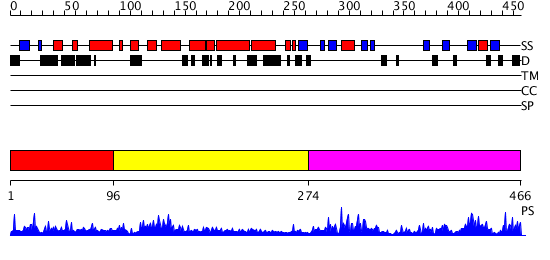
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [96..273] | 14.69897 | Solution Structure of the RNA recognition motif in Peptidyl-prolyl cis-trans isomerase E | |
| 3 | View Details | [274..466] | 59.154902 | Cyclophilin (eukaryotic) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)