GO TERM SUMMARY
|
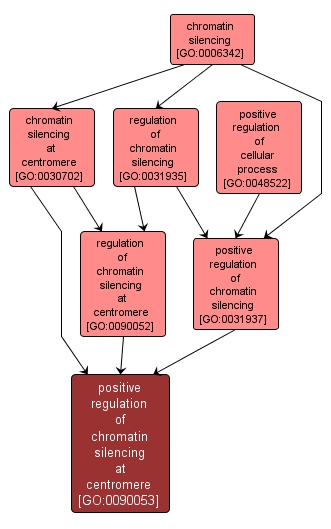
| Name: |
positive regulation of chromatin silencing at centromere |
| Acc: |
GO:0090053 |
| Aspect: |
Biological Process |
| Desc: |
Any process that increases the frequency, rate or extent of chromatin silencing at the centromere. Chromatin silencing at the centromere is the repression of transcription of centromeric DNA by altering the structure of chromatin. |
|

|
INTERACTIVE GO GRAPH
|