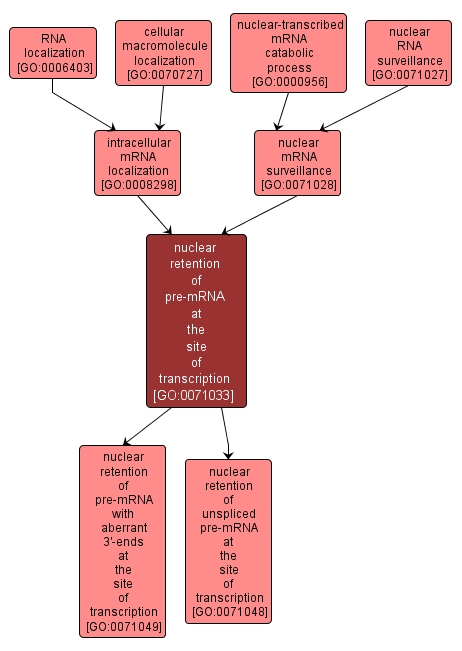
GO TERM SUMMARY
|
| Name: |
nuclear retention of pre-mRNA at the site of transcription |
| Acc: |
GO:0071033 |
| Aspect: |
Biological Process |
| Desc: |
The process involved in retention of aberrant or improperly formed mRNAs, e.g. those that are incorrectly or incompletely spliced or that have incorrectly formed 3'-ends, within the nucleus at the site of transcription. |
|

|
INTERACTIVE GO GRAPH
|