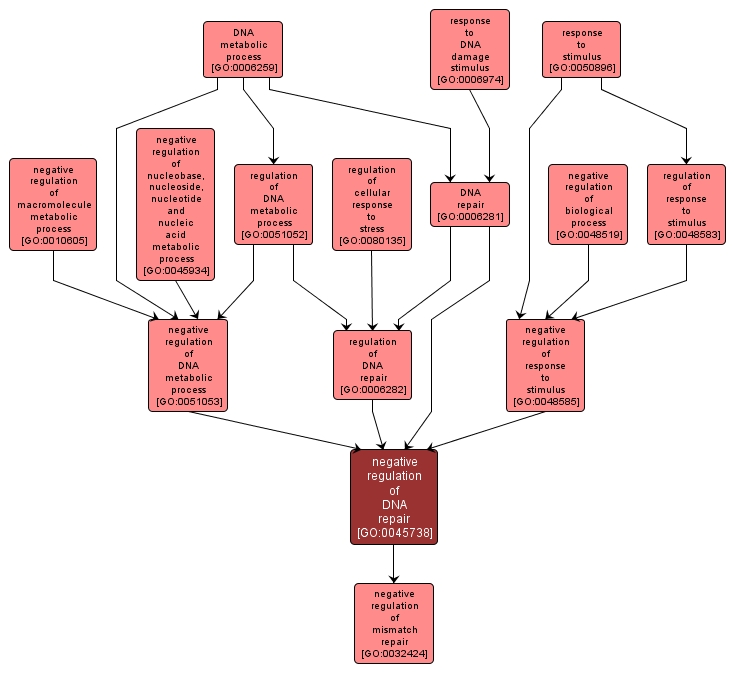
GO TERM SUMMARY
|
| Name: |
negative regulation of DNA repair |
| Acc: |
GO:0045738 |
| Aspect: |
Biological Process |
| Desc: |
Any process that stops, prevents or reduces the frequency, rate or extent of DNA repair. |
Synonyms:
- down-regulation of DNA repair
- down regulation of DNA repair
- downregulation of DNA repair
- inhibition of DNA repair
|
|

|
INTERACTIVE GO GRAPH
|