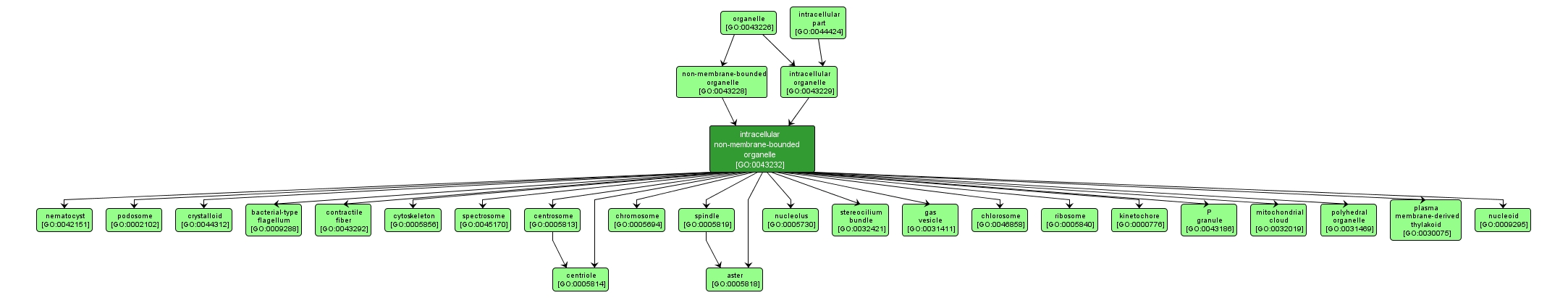
GO TERM SUMMARY
|
| Name: |
intracellular non-membrane-bounded organelle |
| Acc: |
GO:0043232 |
| Aspect: |
Cellular Component |
| Desc: |
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes. |
Synonyms:
- intracellular non-membrane-enclosed organelle
|
|

|
INTERACTIVE GO GRAPH
|