GO TERM SUMMARY
|
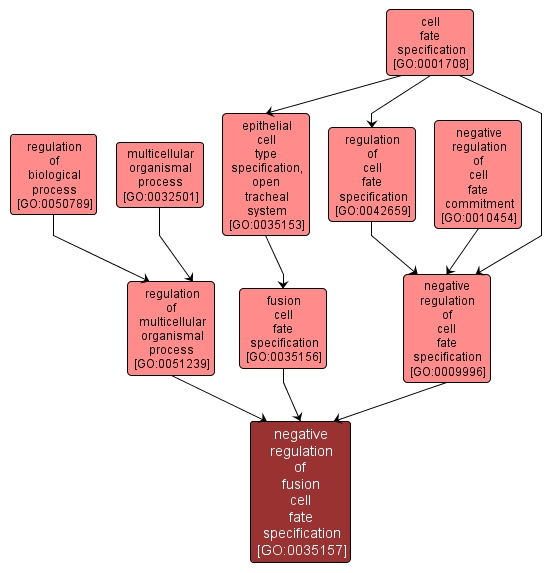
| Name: |
negative regulation of fusion cell fate specification |
| Acc: |
GO:0035157 |
| Aspect: |
Biological Process |
| Desc: |
Any process that restricts, stops or prevents a cell from adopting a fusion cell fate. Once the terminal and fusion fates have been correctly induced, inhibitory feedback loops prevent the remaining branch cells from assuming similar fates. |
Synonyms:
- downregulation of fusion cell fate specification
- inhibition of fusion cell fate specification
- down-regulation of fusion cell fate specification
- down regulation of fusion cell fate specification
|
|

|
INTERACTIVE GO GRAPH
|