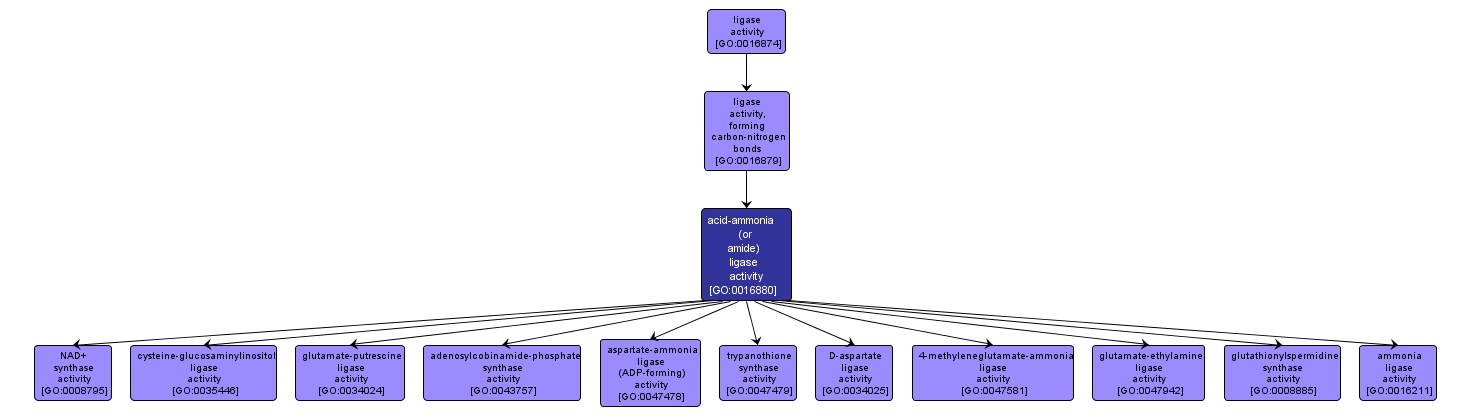
GO TERM SUMMARY
|
| Name: |
acid-ammonia (or amide) ligase activity |
| Acc: |
GO:0016880 |
| Aspect: |
Molecular Function |
| Desc: |
Catalysis of the ligation of an acid to ammonia (NH3) or an amide via a carbon-nitrogen bond with concomitant breakage of a diphosphate linkage, usually in a nucleoside triphosphate. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|