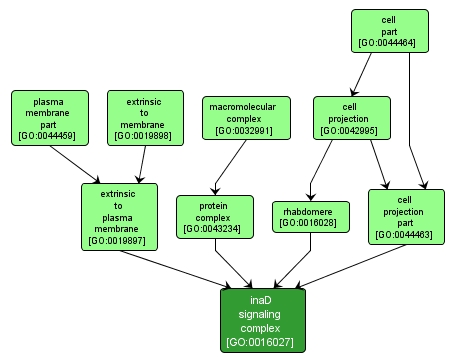
GO TERM SUMMARY
|
| Name: |
inaD signaling complex |
| Acc: |
GO:0016027 |
| Aspect: |
Cellular Component |
| Desc: |
A complex of proteins that are involved in phototransduction and attached to the transient receptor potential (TRP) channel. The protein connections are mediated through inaD. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|