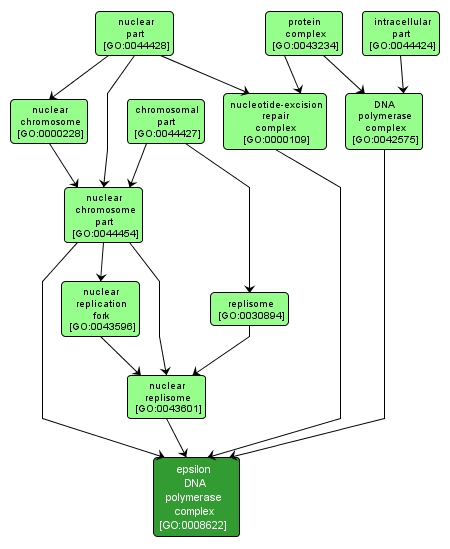
| Desc: |
A heterotetrameric DNA polymerase complex that catalyzes processive DNA synthesis in the absence of PCNA, but is further stimulated in the presence of PCNA. The complex contains a large catalytic subunit and three small subunits, and is best characterized in Saccharomyces, in which the subunits are named Pol2p, Dpb2p, Dpb3p, and Dpb4p. Some evidence suggests that DNA polymerase epsilon is the leading strand polymerase; it is also involved in nucleotide-excision repair and mismatch repair. |