GO TERM SUMMARY
|
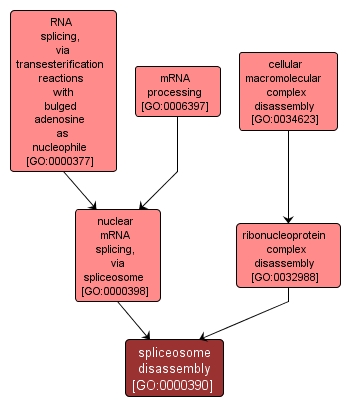
| Name: |
spliceosome disassembly |
| Acc: |
GO:0000390 |
| Aspect: |
Biological Process |
| Desc: |
Disassembly of the spliceosome with the ATP-dependent release of the product RNAs, one of which is composed of the joined exons. In cis splicing, the other product is the excised sequence, often a single intron, in a lariat structure. |
Synonyms:
- spliceosome complex disassembly
- U2-type spliceosome disassembly
- GO:0000392
- GO:0000391
- U12-type spliceosome disassembly
|
|

|
INTERACTIVE GO GRAPH
|